Fig. 3.
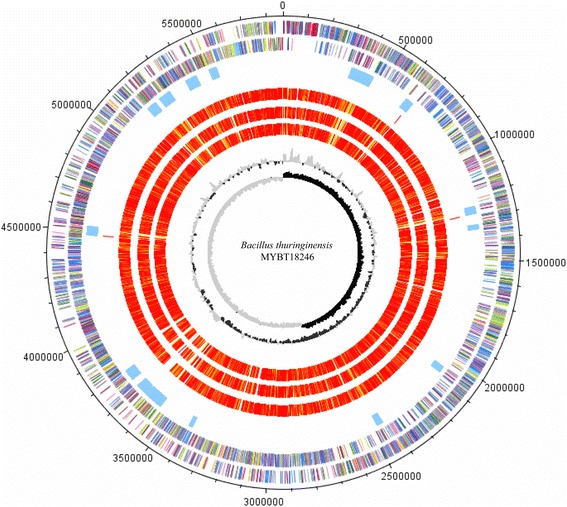
Circular visualization of the genome comparison of 10.1601/nm.5000 MYBT18246 with 3 other sequenced 10.1601/nm.5000 strains. The tracks from the outside represent: (track 1–2) Genes encoded by the leading and lagging strand of 10.1601/nm.5000 MYBT18246 marked in COG colors [93]; (track 3) putative prophage regions, identified with PHAST in blue [76], (track 4) identified cry toxin genes in red; (track 5–7) orthologs for the genomes of 10.1601/nm.5000 YBT-1518 (CP005935.1), 10.1601/nm.5000 CT-43 (CP001907.1), 10.1601/nm.5000 Bt407 (CP003889.1) illustrated in red to light yellow, singletons in grey (grey: <1e−20; light yellow: 1e−21–1e−50; gold: 1e−51–1e−90; light orange: 1e−91–1e−100; orange: 1e−101–1e−120; red: >1e−121 (track 7) % GC plot (track 8), GC skew [(GC)/(G + C)]. Visualization was done with DNAPlotter [94]
