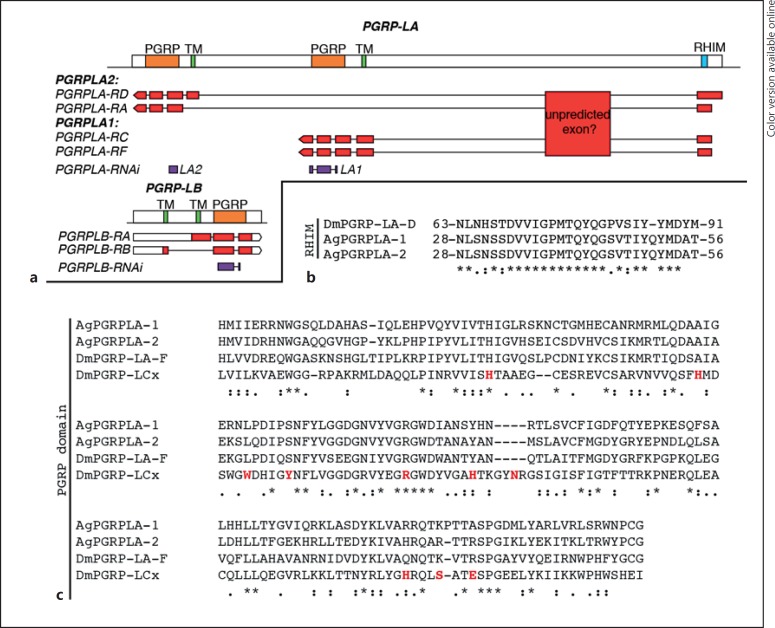
Fig. 4.
Map of PGRPLA and PGRPLB and sequence features of PGRPLA. a Map of the PGRPLA and PGRPLB gene loci. Predicted PGRP domains are shown in orange, predicted transmembrane domains (TM) in green and the predicted RHIM in turquoise (see online version for colors). The map of each VectorBase-predicted transcript is shown below each gene, including untranslated regions in white and coding DNA sequences in red. According to RNA-Seq data [14, 15, 16], a transcribed region that may participate in the sequence of PGRPLA transcripts is also indicated as “unpredicted exon.” dsRNA-targeted regions are shown in purple. b, c Sequence alignments of PGRPLA RHIM (b) and PGRP domain (c) with their homologous sequences in Drosophila PGRPLA (b, c) and PGRPLCx (c). In Drosophila, there is no single isoform encompassing both an RHIM and a PGRP domain, hence the sequence of PGRPLAD isoform is shown for the RHIM and the sequence of PGRPLAF isoform for the PGRP domain. c Amino acids shown in red are in direct contact with the peptidoglycan monomer “TCT” (tracheal cytotoxin) [18]. Sequence alignments were performed with ClustalW [19] and manually curated. *, positions with fully conserved residues;:, positions with conservation of “strong” groups (STA, NEQK, NHQK, NDEQ, QHRK, MILV, MILF, HY, FYW); ·, positions with conservation of “weaker” groups (CSA, ATV, SAG, STNK, STPA, SGND, SNDEQK, NDEQHK, NEQHRK, FVLIM, HFY).