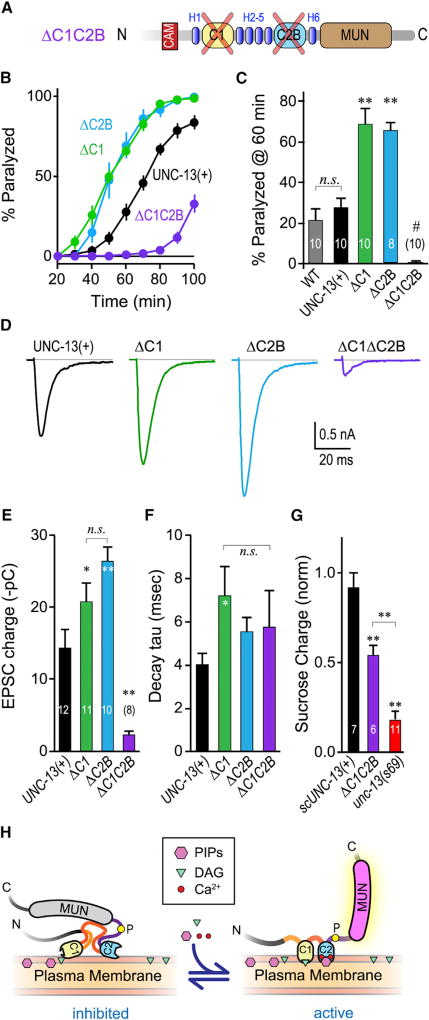
Figure 6. Loss of Both C1 and C2B Nearly Eliminates Calcium-Triggered Fusion.
(A) Schematic of the ΔC1 ΔC2B double domain deletion. The two domains were removed while leaving the surrounding helical regions (H1–H8) intact to minimize global disruption of UNC-13 structure.
(B) Average aldicarb time course for unc-13(s69) rescued with full-length UNC-13 (UNC-13(+); black), ΔC2B (blue), ΔC1 (green), or ΔC1ΔC2B (purple).
(C) Summary of paralysis at 60 min for these four strains as well as WT animals (gray).
(D–F) Average evoked EPSCs (D), average EPSC charge transfer (E), and EPSC decay time constant (F) for the same four strains.
(G) Sucrose-evoked charge (3 s integration; see STAR Methods) for unc-13(s69) (red) alone or rescued with a single-copy full-length UNC-13 (scUNC-13(+); black) or ΔC1ΔC2B (purple). Total charge is normalized to WT.
(H) Model of the C1–C2B protein module in the unliganded (inhibited) state versus the liganded (active) state. The conserved linker region proline is indicated with a yellow circle. Lipid and calcium binding to C1–C2B induces a conformational change that activates a fusogenic function of UNC-13.
Errors bars are mean ± SEM, and experiment number is given within the bars. *p < 0.05, **p < 0.01; #, significantly different from all other strains with p < 0.01; n.s., not significant by ANOVA and Tukey-Kramer. Strains: N2, BC168, JSD805, JSD1038, JSD1039, JSD1064, and JSD1067.