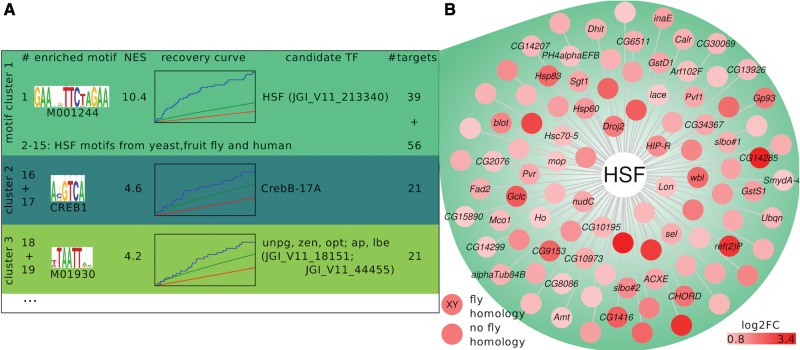
Fig. 2.—
The heat shock factor as a common regulator in heat shock response. We here show Daphnia-cisTarget results for the upregulated genes in a thermal stress experiment (D. pulex exposed for 2 h to a 10 °C temperature increase, whole animal mRNA measured with microarrays). (A) Daphnia-cisTarget returns a heat shock factor (HSF) motif as the top scoring motif (the mouse motif M01244 of the TRANSFAC© Professional database [Matys et al. 2006]), followed by 14 HSF motifs derived from different species such as yeast, fruit fly and human. Those motifs are clustered as highly similar (indicated through same background color). The Daphnia-cisTarget results also show the motif logo (“enriched motif”), the Normalized Enrichment Score (NES), the recovery curve, the D. pulex HSF homologue JGI_V11_213340 derived through sequence homology as candidate transcription factor (TF) and the predicted target genes (“#targets”). The second and third best motif clusters are assigned to a basic-leucine zipper TF CrebB and a range of homeobox TFs, respectively. The remaining 122 recovered motifs that surpass the NES threshold of 2.5 are not shown. (B) The gene regulatory network controlled by the HSF in D. pulex is reconstructed by merging the 95 predicted targets of all HSF motifs recovered by Daphnia-cisTarget. Gene names are derived through homology to D. melanogaster genes; D. pulex genes mapping to the same D. melanogaster gene are numbered (e.g., slbo#1, slbo#2, etc.).