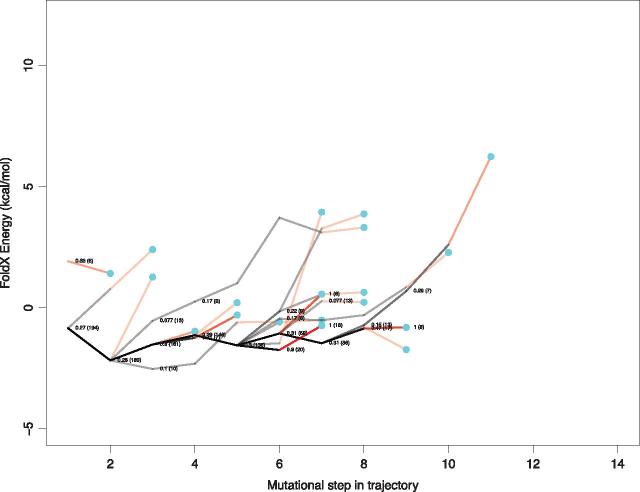
Figure 9.
Energy landscape showing a summary of the cumulative energy changes for all trajectories including those leading to drug resistance in the HIV-1 protease protein. The y-axis represents the predicted difference in energy between each mutant and the wild-type strain (ΔΔG). Each line on the x-axis represents a mutational step between a parent and child node starting from the first descendant of the most recent common ancestor. Each trajectory is terminated at the first mutational step where a drug resistant mutation is detected. The red lines indicate the step in each trajectory just before a resistance mutation is acquired indicated by the blue circles. The probability of a sequence leading to a resistance-associated mutation is shown (total number of sequence passing through that node is in parentheses). The probability of acquisition of a resistance mutation is calculated by dividing the total number of pathways passing through a node that ultimately acquire a resistance mutation by the total number of pathways. For clarity values are omitted if fewer than five sequences pass through a node.