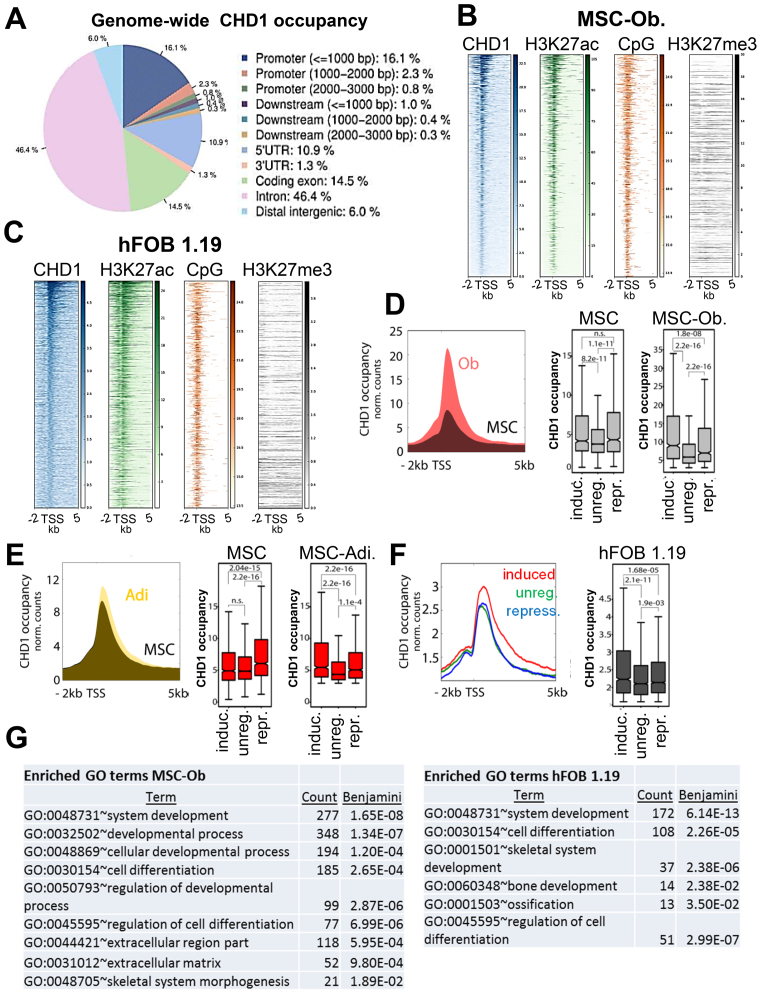
Figure 2.
CHD1 occupancy increases near the TSS of differentiation-induced genes. (A) Relative genome-wide CHD1 occupancy analysis at regulatory elements in osteoblast-differentiated MSC (MSC-Ob.) represented by a pie chart with the color code on the right. Analysis was performed via CEAS. (B, C) Occupancy of H3K27ac, CpG frequency and H3K27me3 relative to CHD1-occupied transcriptional start sites (TSS) ordered from high to low based on CHD1 occupancy in MSC-Ob. (B) and hFOB 1.19 (C). (D) Aggregate plot of CHD1 around the TSS of osteoblast differentiation-induced genes in undifferentiated (black) and differentiated (red) conditions. Quantification of CHD1 occupancy at induced (induc.), unregulated (unreg.) or repressed (repr.) gene groups in undifferentiated (MSC) and differentiated (MSC-Ob) conditions are shown on the right. P-values below 0.05 are shown unless indicated as not significant (n.s.). (E) Aggregate plot of CHD1 as described in D but for adipocyte differentiation-regulated gene groups (Adi.). (F) Aggregate plot of CHD1 around the TSS in hFOB 1.19 at genes induced (red), unregulated (green) or repressed (blue) during osteoblast differentiation. Boxplots on the right show quantitation of CHD1 occupancy with P-values indicated above boxplots. (G) The lists represent significantly enriched gene ontology (GO) terms at high CHD1-occupied genes around the TSS of osteoblast-differentiation induced genes in MSC-Ob. and hFOB 1.19. GO analysis was performed with the DAVID analysis tool with the adjusted P-value (Benjamini) and count of genes within the respective term.