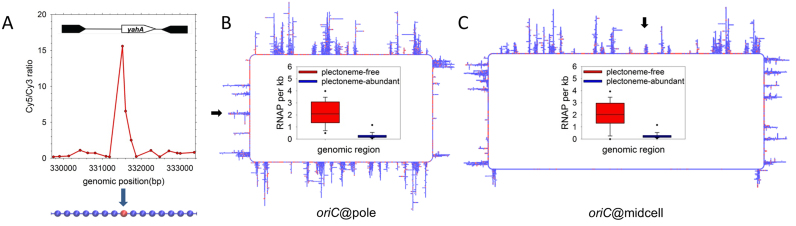
Figure 3.
Design of initial ‘blueprint’ models. (A) Incorporation of ChIP data defining RNAP binding sites (data obtained and figure adapted from (33)). ChIP-determined binding peaks were used to assign identities to beads in the 500 BPB model (lower portion of panel): RNAP beads (beads whose sequence includes a RNAP binding site) appear in red; bulk DNA beads are blue. (B) A representative 500 BPB oriC@pole ‘blueprint’ structure. Regions of active transcription, as indicated by ChIP data, are plectoneme-free regions (PFRs), while the balance of the genome is plectonemic (PARs). oriC is located at the pole of the cell (black arrow on left side of the ‘blueprint’); the inset describes the difference in RNAP density between PFRs and PARs. (C) A representative 500 BPB oriC@midcell ‘blueprint’ structure. The origin is located in the center of the upper arm of the chromosome (black arrow above the ‘blueprint’); the bottom ‘crossing’ region (71) is modeled as plectoneme-free.