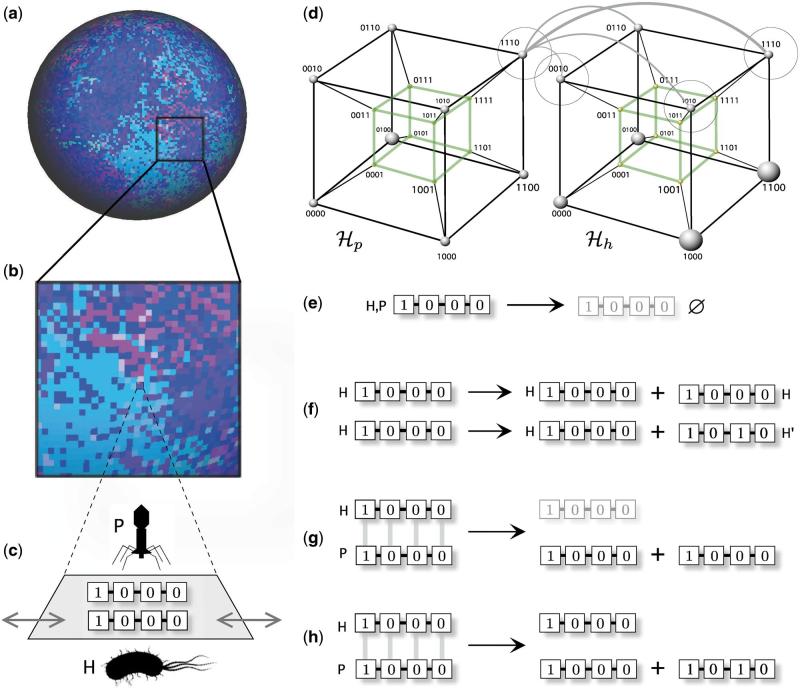
Figure 2.
Coevolution in coupled landscapes associated to a phage-bacteria model based on a digital genomes representation. Each individual is described by means of a digital genome of length ν. The model dynamics is defined on a two-dimensional surface (a) where the color of each site indicates (in this case) the presence of different strings. In (b), an expanded area shows the presence of local patchiness where each site (c) can be occupied by one string of each class. Strings can move randomly to neighboring free sites, as indicated by the grey arrows. From the point of view of the genotype space, we have two coupled sequence hypercubes (here ν = 4) where similarity between phage and bacteria recognition sequences (i.e. the matching allele model or recognition, determines the probability of interaction. Two different hypercubes are shown, one for phages (left) and another (right) for bacteria. One given virus (like 1110) will be able to interact with those bacteria whose genome is closer (the probability of this interaction is indicated by weighted gray lines). The basic rules used in the model are summarized in (e–h). These are: (e) sequence removal (death), (f) replication of host string, which can be accurate H → 2H or inaccurate , for the phage–bacteria interaction.