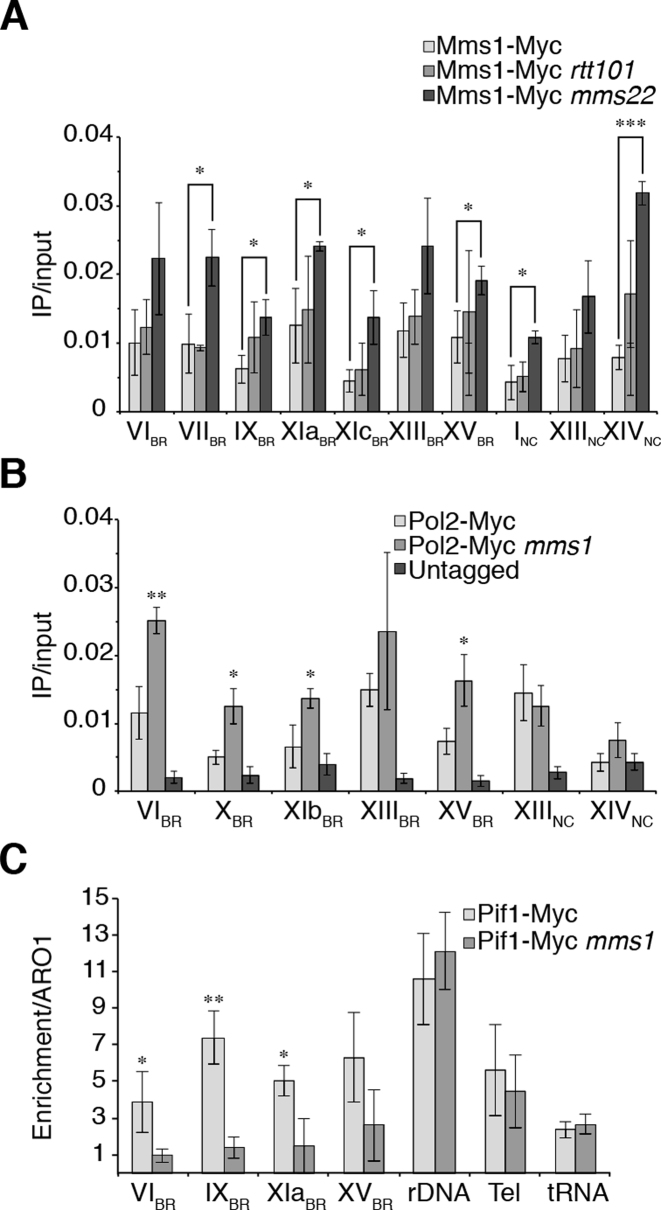
Figure 3.
Mms1 binds independently of Rtt101 and Mms22, supports Pif1 binding at G4 motifs and by this promotes DNA replication. (A) ChIP and qPCR analysis of Mms1-Myc at seven BR and two NC. Binding of Mms1 was monitored in wild type (light), rtt101 (grey) and mms22 (dark) cells. Statistical significance compared to Myc-tagged Mms1 wild type cells. For details on regions see Supplementary data 3. (B) Replication fork progression was analyzed by detected DNA Pol2 binding levels at Mms1 binding sites. ChIP and qPCR of DNA Pol2 binding in wild type and mms1 cells was performed at five BR and two NC. Statistical significance compared to Myc-tagged DNA Pol2 wild type cells. (C) Binding of Pif1 DNA helicase was analyzed at four Mms1 binding regions (BR) in wild type and in mms1 cells. As control for Pif1 binding we used two known Pif1 binding sites, the replication fork barrier at the rDNA (rDNA) (54) and telomere VI-R (tel) (53) as well as one Pif1 independent site (tRNA). As done previously (53) IP/input values are compared to IP/input values of ARO1 where no Pif1 binds. Here, fold enrichment over ARO1 was plotted as mean value ± SD. For all ChIP; N ≥ 3 biological replicates. Statistical significance was determined by Student's t-test. *P < 0.05, **P < 0.01, ***P < 0.001.