Figure 7.
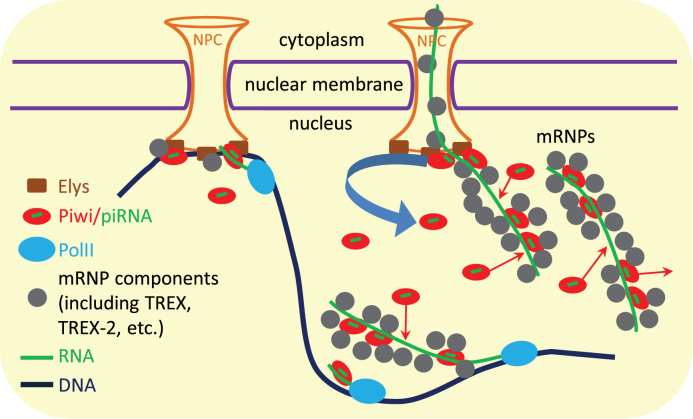
Working model of Piwi circuit in the nucleus. Piwi stochastically interacts with the nascent transcripts of genes and TEs scanning them for the complementarity with loaded piRNAs. While capturing the perfect complementarity, Piwi triggers the transcriptional silencing of TEs. However, while revealing the imperfect complementarity, Piwi remains bound with the transcripts in the mRNPs after their detachment from the sites of transcription and continues to bind and to dissociate from mRNPs in the nucleoplasm. mRNP components may isolate the RNA-bound Piwi molecules from contacts with DNA. Piwi is released from the passing through nuclear pore mRNPs by binding to NPCs (via Xmas-2, Elys or other NPC components). Therefore, different pools of Piwi may exist in the nucleus. The smallest pool is composed from transiently linking to NPCs Piwi molecules, which interact with the attached to NPCs chromatin and, as a result, slightly more efficiently bind and, probably, scan the transcripts of nearby genes and TEs. The released from NPCs, more abundant Piwi fraction moves by Brownian motion to the chromatin positioned inside the nucleoplasm, where Piwi may scan the nascent transcripts and may bind with mRNPs. Finally, the most abundant Piwi fraction is represented by already bound to mRNPs Piwi.