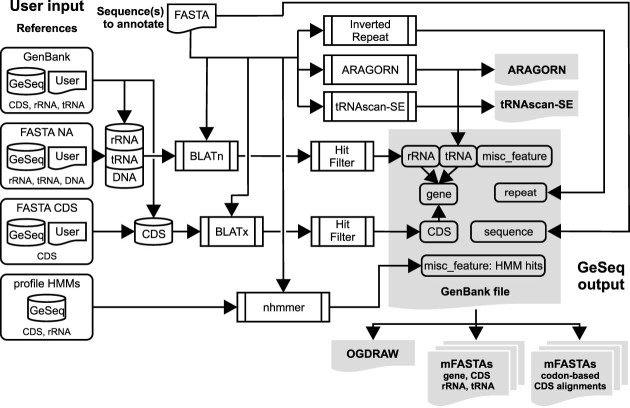
Figure 2.
GeSeq annotation pipeline. The user provides nucleic acid FASTA sequence(s) for annotation and selects or provides reference nucleic acid sequences in GenBank or FASTA format (‘User Input’). Based on the selected or uploaded reference sequences (‘References’), GeSeq builds a non-protein-coding (rRNA, tRNA and DNA) and a protein-coding (CDS) BLAT database, carries out standard (‘BLATn’) and translated BLAT (‘BLATx’) searches, respectively, and filters the hits (‘Hit Filter’). GeSeq annotates from the filtered hits the classes rRNA, tRNA, CDS and gene (‘gene entries’). ‘Gene entries’ result from tRNA, rRNA and CDS hits, and include introns (if present). DNA hits are annotated as ‘misc_features’ (as shown here) or, alternatively, as ‘primer_bind’ if invoked by the user (see text for details). In addition, the user can activate an nhmmer search by selecting profile HMMs of CDS and rRNA sequences (currently chloroplast only) as references. All profile HMM hits are annotated as misc_features to support manual curation. Optionally, the user can invoke ARAGORN or tRNAscan-SE for de novo annotation of tRNAs and a self-BLATn search for the detection of the inverted repeat (IR) pair typically found in chloroplast genomes. The minimum GeSeq output (all output files are labeled in gray) is a GenBank file that contains all annotations and its interpretation by OGDRAW for a quick evaluation. Additionally, the user can choose additional optional outputs, including separate multi-FASTA files (‘mFASTAs’) containing the annotated sequences belonging to the classes gene, CDS, rRNA and tRNA. If several sequences were uploaded for annotation in the same job, also combined mFASTAs for all annotated sequences of the four classes are offered for download and optionally, codon-based alignments can be produced for all annotated CDS sequences with or without the selected or uploaded GenBank references.