Figure 1.
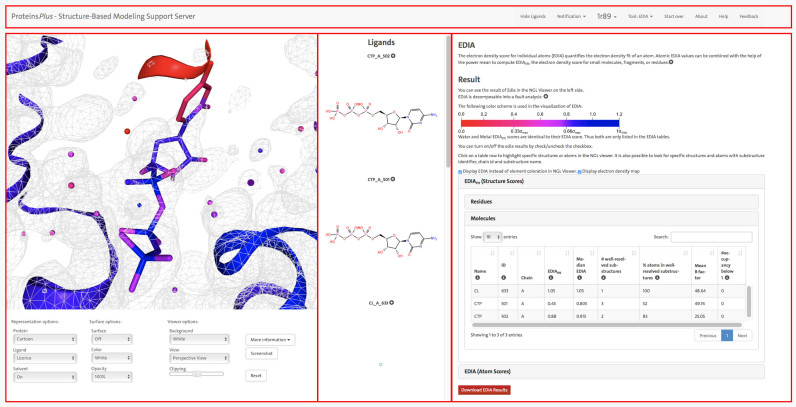
EDIA analysis for the crystal structure of an archaeal class I CCA-adding enzyme in complex with cytidine-5΄-triphosphate (CTP) (PDB ID: 1R89 (10)). This figure demonstrates how the ProteinsPlus web server can be used to assess the quality of a protein structure and analyze potential uncertainties in the structure. The panel on the right side shows the results from the EDIA calculation along with a short description of the quality measure. The detailed results for the EDIAm (structure score) for molecular substructures are displayed. For CTP 501 A, the EDIAm score is very low, indicating possible uncertainties in the structure. The binding site of this CTP molecule is shown in the left panel in the NGL web viewer, allowing a detailed visual inspection. All atoms in the structure are colored according to their individual EDIA score (as explained in the right panel). Additionally, the electron density map (2fo-fc) at 1 σ is displayed. It is clearly recognizable that most atoms in the cytosine moiety receive very low EDIA scores. This is consistent with the observation that around these atoms no electron density is observed at 1σ. The figure also highlights the menu bar at the top and all three panels with red rectangles, the NGL viewer with the control panel on the left, the ligand panel with structure diagrams in the middle and the tool panel with the result page of EDIA at the right.