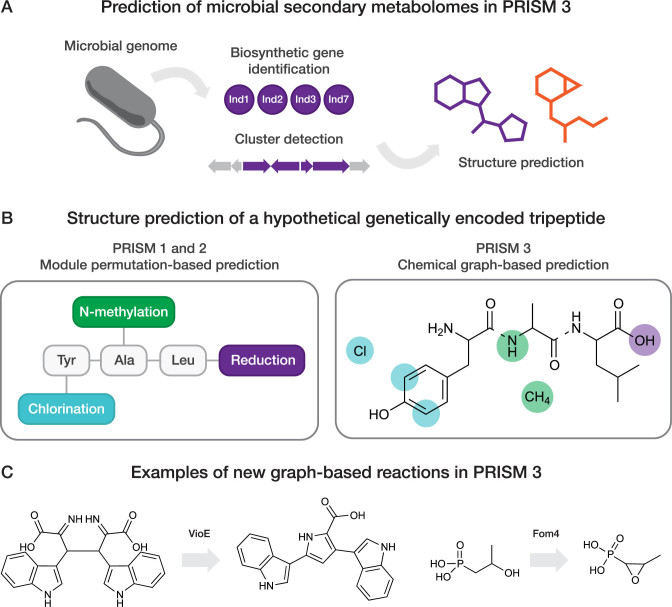
Figure 1.
(A) Schematic overview of microbial secondary metabolome prediction in PRISM 3. Following ORF detection in a microbial genome sequence, protein sequences are analyzed and clustered using a library of hidden Markov models for secondary metabolite biosynthesis genes. Identified biosynthetic information is subsequently leveraged for combinatorial prediction of secondary metabolite chemical structures. (B) Overview of chemical graph-based secondary metabolite structure prediction in PRISM 3. Modeling a natural product as a chemical graph, rather than a linear permutation of monomers, facilitates manipulation of the predicted structure at the level of individual atoms or bonds rather than at the level of the monomers. In PRISM 3, individual sets of atoms, rather than individual sets of modules, are tagged as potential sites of tailoring reactions before combinatorialization. Linkages between residues within the same subgraph are indicated as dashed lines. (C) Examples of new virtual reactions facilitated by graph-based structure prediction in PRISM 3.