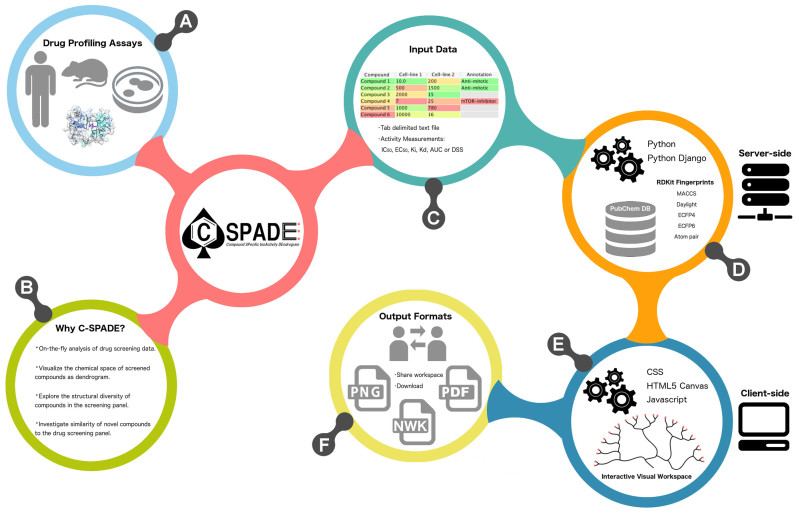
Figure 1.
An infographic illustration of C-SPADE web-tool and its functionalities. (A) Various types of biological drug screening assays (biochemical, cell-based, cell-free or target-based assays) can be analyzed. (B) The key functionalities implemented in C-SPADE. (C) The input consists of a tab-delimited text file of drug screening data, with compound names and bioactivity values, where a wide range of activity measurements are supported. (D) On the server-side, the compound name is used to retrieve the compound structural information from the PubChem database and various compound features are calculated using the RDKit package. (E) The client-side options provide an interactive interface to visualize and customize the compound clusters. (F) The output visualization from C-SPADE can be either shared with collaborators or downloaded as publication-quality images or as a Newick tree file.