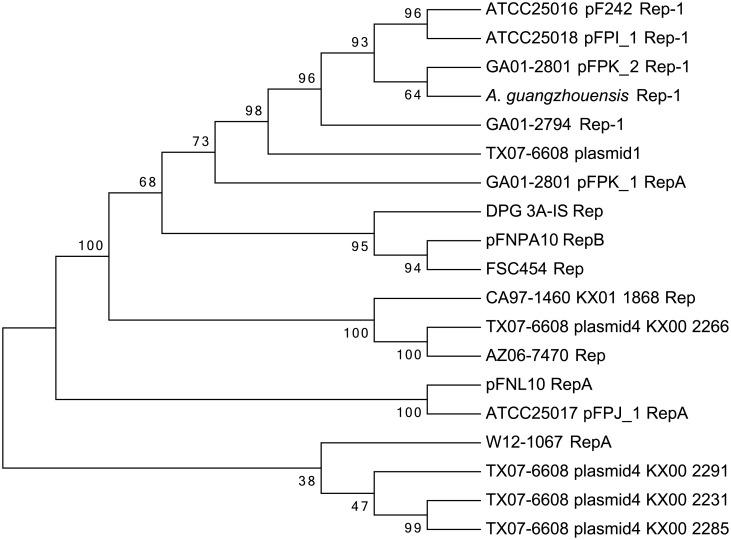
Fig 4. Phylogenetic analysis of putative Rep protein sequences.
Evolutionary history was inferred by using the Maximum Likelihood method based on the JTT matrix-based model. The bootstrap consensus tree inferred from 500 replicates represents the evolutionary history of the taxa analyzed. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (500 replicates) are shown next to the branches. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using a JTT model, and then selecting the topology with superior log likelihood value. This analysis involved 19 amino acid sequences. There were a total of 551 positions in the final dataset. The Rep protein sequences KX00_2285 and KX00_2291, from TX07-6608 plasmid 4, were partial sequences.