Fig 3. Density-associated mutation-rate plasticity (DAMP) in strains of E. coli with deficiencies in various DNA repair and other systems.
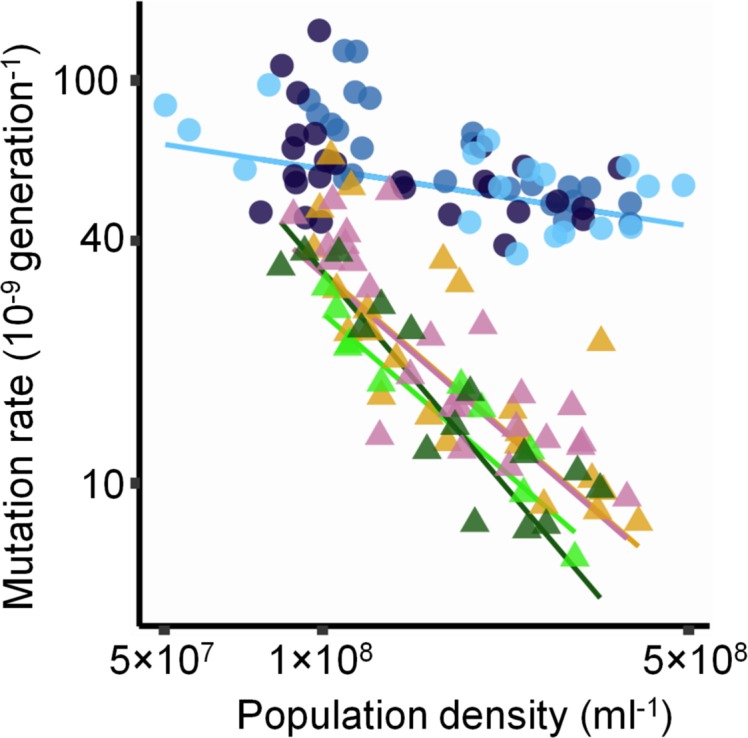
Mutation rate in strains from the Keio collection [24] deleted for genes encoding deoxyadenosine methylase (Δdam, N = 23, yellow triangles), error-prone DNA polymerase Pol IV (ΔdinB, N = 25, pink triangles), methionine transporter MetI (ΔmetI, N = 10, light green triangles), endonuclease VIII (Δnei, N = 15, dark green triangles), and each protein of methyl-directed DNA mismatch repair (MMR), which comprises MutS (ΔmutS, N = 28, light blue circles) that binds mismatched DNA and MutL (ΔmutL, N = 27, dark blue circles) that interacts with MutS to activate endonuclease MutH (ΔmutH, N = 24, medium blue circles). All lines result from Model S-VII in S1 Text. Wald tests that average slope for Δdam, ΔdinB, and ΔmetI is 0: t92 = 17; P = 4.2×10−30, that average slope for Δnei is 0: t91 = 13; P = 6.0×10−22, and that average slope for MMR strains is 0: t91 = 4.3; P = 5.1×10−5. See S2 Table for further strain details. Densities estimated by ATP-based luminescence assay. As in Fig 2, triangles and circles indicate mutation rates to rifampicin and nalidixic acid resistance, respectively. Note the logarithmic axes. Raw data are available in S1 Data.
