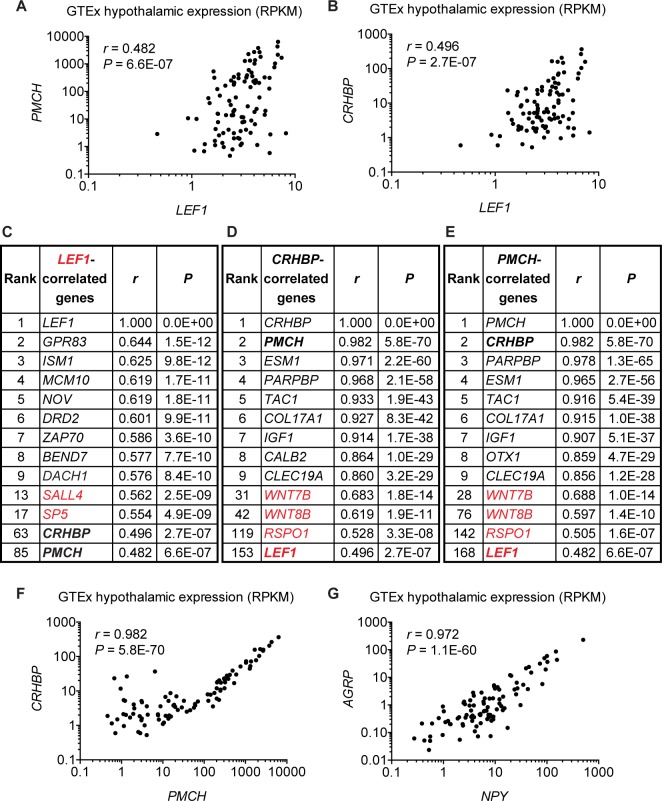
Fig 7. Correlation analysis in the human hypothalamus.
(A-G) Pearson correlations for hypothalamic gene expression among 96 postmortem human samples obtained from the Genotype-Tissue Expression (GTEx) project [39]. All the Pearson’s r and P values were calculated between 2 genes, and displayed in the graphs or tables after sorting by r values. Correlation expression profiles are shown for gene pairs Pro-melanin concentrating hormone (PMCH) versus LEF1 (A), Corticotropin-releasing hormone binding protein (CRHBP) versus LEF1 (B), CRHBP versus PMCH (F), and Agouti-related protein (AGRP) versus Neuropeptide Y (NPY) (G), with reads per kilobase of transcript per million mapped reads (RPKM) at log10 scale used on both axes. Note that 1 data point (NPY: 0.1395; AGRP: 0) was not included in (G) due to the inability of plotting a 0 value on the logarithmic axis. Three tables of correlated genes for LEF1 (C), CRHBP (D), and PMCH (E) list the top 9 positively correlated genes plus selected genes, including those involved in canonical Wnt signaling labeled in red. See the full list in S8 Table.