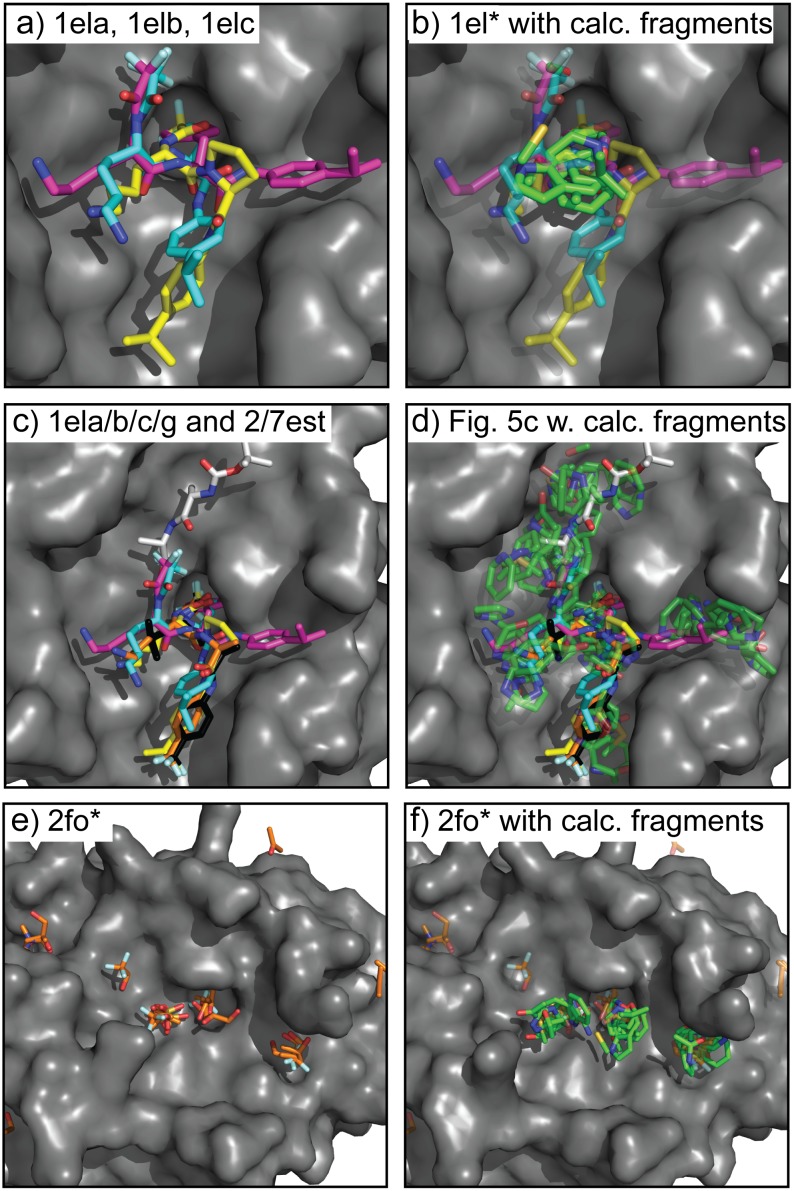
Fig 9. Hot spots on elastase.
Overlay of 3 different (a) elastase co-crystal structures 1ela (yellow), 1elb (cyan), and 1elc (magenta). Beginning in 1976 and continuing until 1994 all of the elastase co-crystals had the ligand binding in the same mode as 1ela. The 1elb and 1elc structures solved in 1994 revealed new binding subsites. The highest affinity (b) SACP fragment clusters (green carbons) with water exclusion are at the site where the 3 ligands overlap. This result is the same for simulations using the 1ela, 1elb, and 1elc structures. Nine co-crystal (c) structures (2fo9, 2foa, 2fob, 2foc, 2fod, 2foe, 2fof, 2fog, 2foh) are needed to map out the entire binding site in elastase. SACP fragment binding predictions using the top three highest interactions (d) cover the subsites from all 9 co-crystals, indicating that these sites were long undiscovered because that have lower affinity for the ligands. Importantly, experimental X-ray fragment-based soaking experiments (e) done in the Ringe Lab did reveal the lower affinity hard-to-find subsites. Using this crystal structure as input into SACP, the simulations (f) also find all of the subsites.