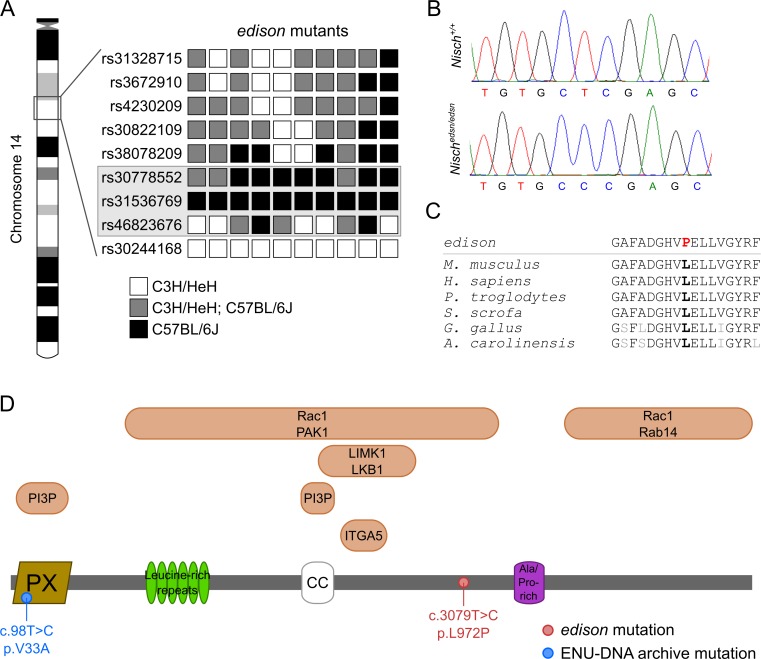
Fig 1. The Nisch gene is mutated in edison mice.
(A) SNP mapping of 10 edsn mutants identified an approximately 9 Mb interval on chromosome 14 delineated by marker rs30778552 and rs46823676. The grayed box indicates the chromosomal interval bearing the edsn mutation. (B) Sequence analysis of the Nisch locus in Nisch+/+ and Nischedsn/edsn DNA. A c.3079T>C transition is detected in Nischedsn/edsn mutants that is not present in Nisch+/+ DNA. (C) Conservation of the mutated leucine residue across species. Mus musculus, ENSMUSG00000021910; Homo sapiens, ENSG00000010322; Pan troglodytes, ENSPTRG00000015001; Sus scrofa, ENSSSCG00000011442; Gallus gallus, ENSGALG00000043825; Anolis carolinensis, ENSACAG00000006939. (D) Schematic of the full length NISCH peptide (1593 amino acids). The molecule consists of a phox-homology (PX) domain, six leucine-rich repeats, a coiled-coil (CC) domain and an alanine/proline-rich region. Both the PX and CC domains of Nischarin are essential for endosomal targeting and interaction with phosphatidylinositol 3-phosphate (PI3P) in PI3P-enriched endosomes. Amino acids 709–807 of Nischarin interact with the integrin α5 (ITGA5) cytoplasmic tail. Both LIMK1 and LKB1 interact with positions 661–869 of Nischarin. Residues 246–1047 of Nischarin interact with PAK1. Rab14 interacts with amino acids 1190–1593. Finally, Rac1 interacts with two regions of Nischarin, amino acids 246–1047 and 1190–1593. The positions of the Nischedsn and NischV33A mutations are also indicated.