Extended Data Figure 9. Szt2 is essential for mTORC1 inactivation under nutrient deprivation conditions in mice and MEFs.
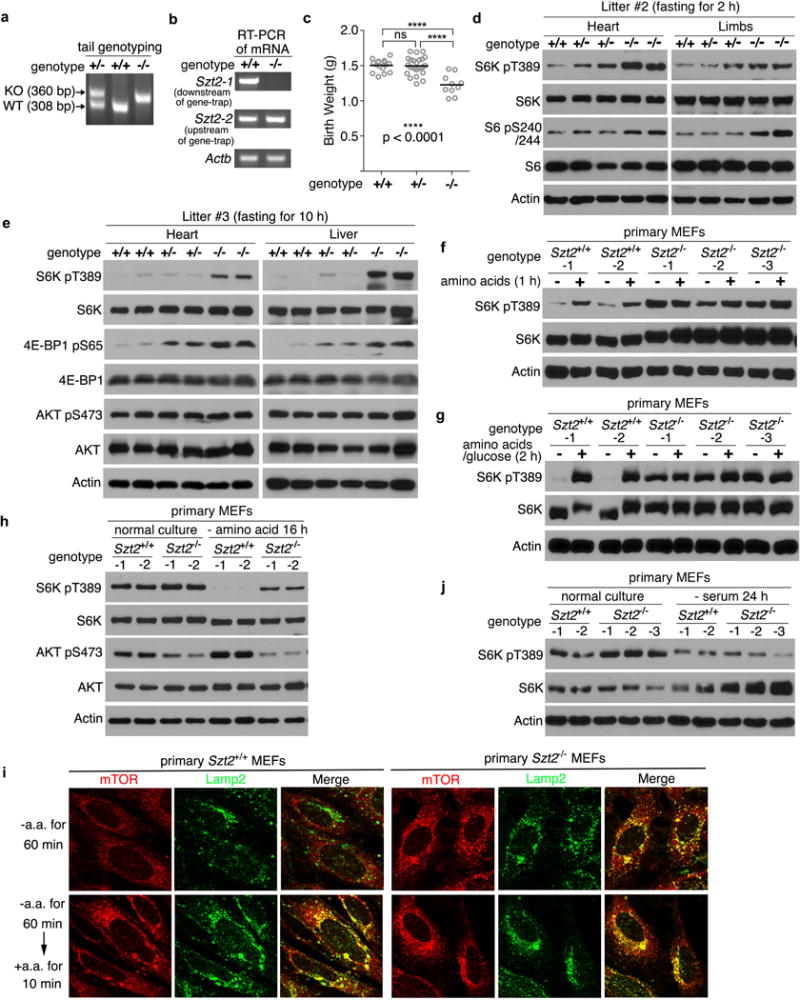
a, PCR genotyping of Szt2+/+, Szt2+/−, and Szt2−/− mice. The expected sizes of wild-type (WT) or Szt2 mutant (KO) alleles are indicated by arrows. b, RT-PCR analysis of mRNA of Szt2 in Szt2+/+ and Szt2−/− MEFs. Primer pair 1 (Szt2-1) detects a region downstream of the gene-trap, and primer pair 2 (Szt2-2) detects a region upstream of the gene-trap. c, The birth weight of neonates from the indicated genotypes are shown. Data represent mean ± SD, n =10 ~ 25, two-tailed unpaired t-test. ns, not significant. d, e, Neonates were fasted for 2 h (d) or 10 h (e). Total cell lysates prepared from the indicated organs were analyzed by immunoblotting. f, g, Szt2+/+ and Szt2−/− MEFs (< 4 passages) were deprived of amino acids for 1 h (f), or amino acid plus glucose for 2h (g) and stimulated with amino acids (f), or amino acid plus glucose (g) for 10 min when indicated. Total cell lysates were analyzed by immunoblotting. h, Szt2+/+ and Szt2−/− MEFs (< 4 passages) were cultured in complete medium or starved with amino acids for 16 h and analyzed as in (f). i, Szt2+/+ and Szt2−/− MEFs (< 4 passages) were deprived of amino acids for 1 h and stimulated with amino acids for 10 min when indicated. The localization of mTOR and Lamp2 was determined by immunostaining. j, Szt2+/+ and Szt2−/− MEFs (< 4 passages) were cultured in complete medium or serum-starved for 24 h and analyzed as in (f). Data (f – j) are representatives of two independent experiments.
