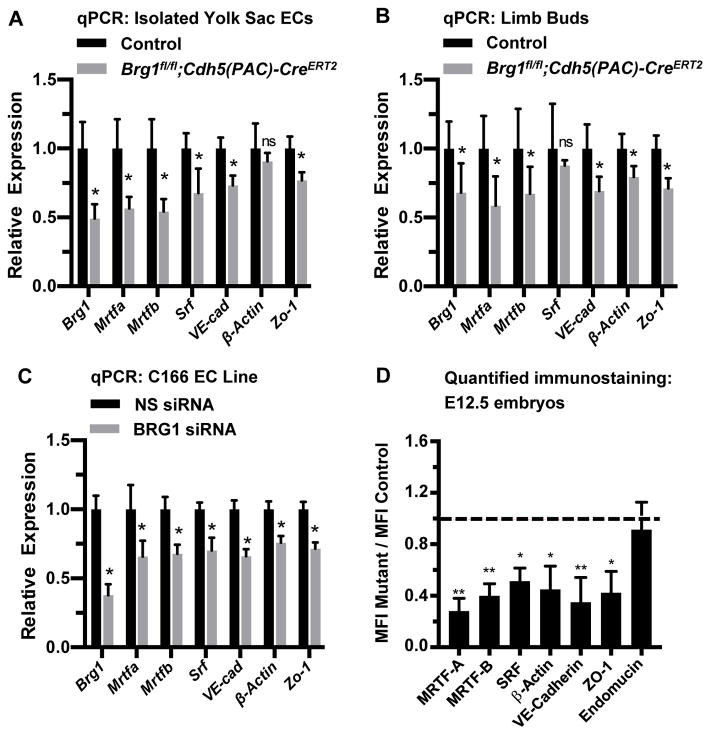
Figure 3. Srf, Mrtfa, Mrtfb, and endothelial cell SRF target genes are downregulated in BRG1-depleted endothelial cells and tissues.
(A–C): Total RNA was collected from freshly isolated yolk sac endothelial cells (ECs; A) or from limb buds (B) of E12.5 control (Brg1fl/fl) and Brg1fl/fl;Cdh5(PAC)-CreERT2 embryos for transcript analysis of Brg1, Srf, SRF co-factors (Mrtfa and Mrtfb), and known endothelial cell SRF target genes (VE-Cadherin, β-actin, and Zo-1) by qPCR. The same transcripts were likewise analyzed in the murine C166 yolk sac EC line following transfection with nonspecific (NS) or BRG1 siRNA oligos for 24 hr (C). For A–C, data were normalized to the relative expression of control samples. Error bars represent S.D. of results from at least three independent experiments or from littermate control and mutant embryos from three different litters. (D) Immunostaining for the vascular marker endomucin and for the gene products of the transcripts analyzed in A–C was performed on sections of E12.5 littermate control and Brg1fl/fl;Cdh5(PAC)-CreERT2 embryos and was quantified (representative immunostaining images used for quantification are shown in Figures VII and VIII in the online-only Data Supplement). Average mean fluorescent intensities (MFI) of mutant capillaries normalized to MFI in control capillaries (dotted line) are shown. Data were compiled from neck and limb bud capillary vessels from 3 images taken from 3 different sets of littermate control and mutant embryos; error bars represent S.D. All statistical calculations were performed using a two-tailed Student’s t test (*, P<0.05; **, P<0.01; ns=not significant).