Figure 5.
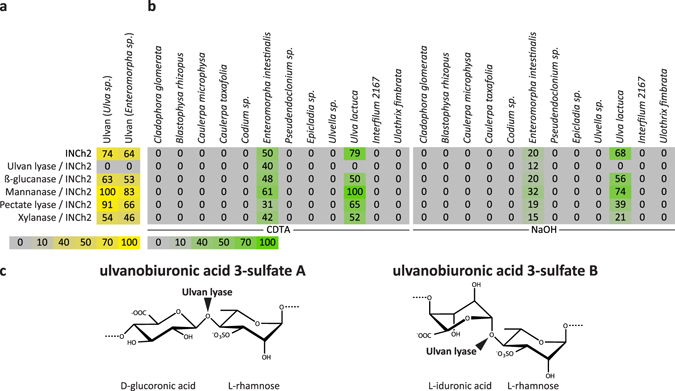
INCh2 characterisation using a microarray populated with ulvan polysaccharides and extracted cell wall material of diverse Ulvophycea species. (a) Heatmap showing INCh2 binding to a carbohydrate microarray populated with ulvan polysaccharides from the species Ulva sp. and Enteromorpha sp. and enzymatic epitope deletion. INCh2 binding was abolished by pre-treatment of arrays with ulvan lyase but not by any control enzymes. INCh2 binding was abolished by pre-treatment of arrays with ulvan lyase but not by control enzymes. (b) Heatmap showing INCh2 binding to a carbohydrate microarray populated with extracted cell wall material of diverse Ulvophycea species and enzymatic epitope deletion. The heatmap shows the relative abundance of cell wall components as extracted using CDTA and NaOH. The same amount of cell wall material (alcohol insoluble residue) was used for each sample. Note that INCh2 bound to Ulva lactuca and Enteromorpha intestinalis containing ulvan, but that INCh2 binding was only abolished by pre-treatment of arrays with ulvan lyase on Ulva lactuca. (c) Diagram showing the cleavage sites of the ulvan degrading enzyme used in the major disaccharide units found in ulvan. Colour intensity in the heatmap is correlated to mean spot signals. The highest signal in each dataset was set to 100, and all other values were normalized accordingly as indicated by the colour scale bar. A low-end cut-off value of 5 was imposed.
