Figure 1.
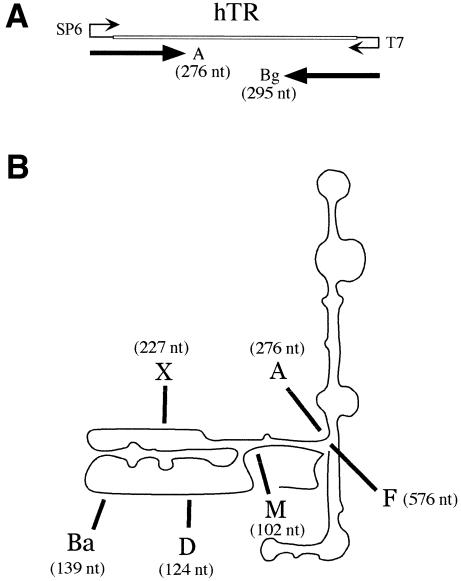
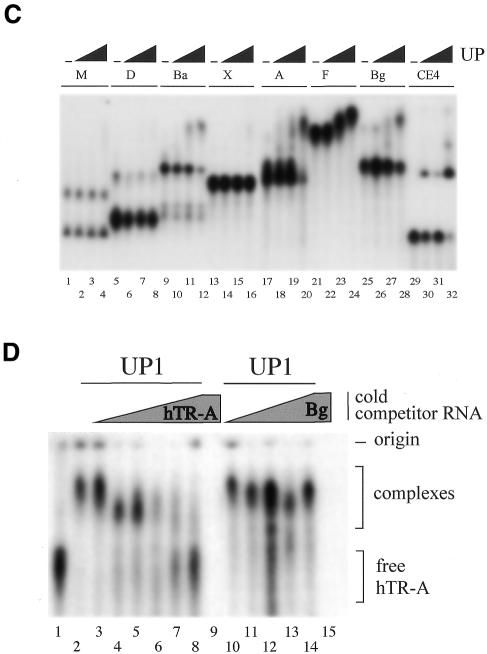
UP1 binds specifically to hTR. (A) Diagram of the hTR portion (451 bp) of the pGem-T plasmid with the transcription start sites of the SP6 and T7 RNA polymerases. hTR-A RNA is 276 nt, the first 68 nt of which are plasmid sequences. Bg RNA is a control RNA derived from the complementary strand. (B) A schematic representation of hTR is shown and is based on the report by Chen et al. (34). The 3′ end of each transcript used in (C) is indicated and corresponds to the position used for plasmid linearization and run-off transcription with SP6 RNA polymerase. The lengths of the products obtained after transcription from the pGEM-T plasmid are indicated in parenthesis and include the 5′ portion derived from plasmid sequence. (C) The binding of UP1 to various portions of hTR. Mixtures containing recombinant UP1 and portions of hTR or control RNAs were fractionated in a 5% native polyacrylamide gel. Each set contains the RNA alone (1 fmol) and increasing amounts of UP1 protein (2.5, 5 and 10 pmol). This gel-shift assay was performed in the presence of heparin at a final concentration of 2 µg/µl. (D) UP1 binding in the presence of competitor RNAs. UP1 (5 pmol) and the 32P-labeled hTR-A RNA (1 fmol) were incubated in the presence of increasing amounts of competitor hTR-A RNA (1, 50, 100, 250, 500 and 1000 fmol) or Bg RNA (1, 50, 100, 500 and 1000 fmol). Because the unlabeled RNAs were synthesized in the presence of residual amounts of [32P]-UTP to facilitate purification and quantification, aliquots of ‘cold’ hTR-A and Bg RNAs alone were loaded as controls (lanes 9 and 15, respectively).