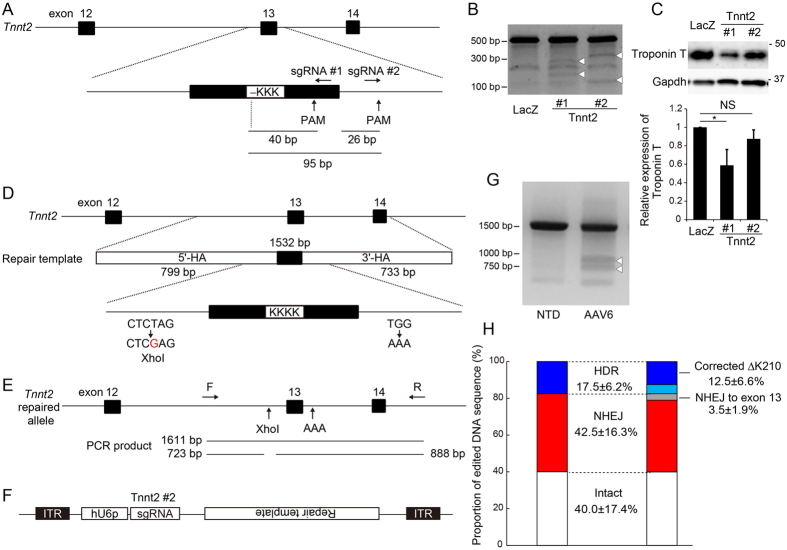
Figure 4.
HDR-mediated genome editing targeting a pathological deletion mutation in dilated cardiomyopathy (DCM) model mice. (A) SgRNA-targeted site around exon 13 in the mouse Tnnt2 gene. Distances from ΔK210 or 3′-terminal of exon 13 to each PAM sequence are shown. (B) Neonatal cardiomyocytes isolated from Cas9 knock-in mice were transfected with sgRNA against LacZ (control), or sgRNA #1 or #2 targeting Tnnt2. Forty-eight hours after transfection, genomic DNA was extracted, and cleavage activities were evaluated using the Cel-I assay. White arrowheads indicate cleaved PCR products. (C) Whole-cell lysates were extracted from the cells treated as in (B). Troponin T and Gapdh were detected by western blot. Expression level of Troponin T was normalized against the corresponding level of Gapdh (n = 3, means ± SD, *p < 0.05 vs LacZ). (D) Design of HDR repair template consisting of 799-bp 5′-terminal and 733-bp 3′-terminal homology arms (5′-HA and 3′-HA) corresponding to genomic sequence around exon 13 of Tnnt2. XhoI recognition site and PAM mutation (TGG to AAA) were introduced into both arms. (E) Detection of HDR by XhoI digestion of PCR products generated from primers inside (F) and outside (R) the homology arms. Expected lengths of PCR products are shown. (F) Design of AAV vector. hU6 promoter (hU6p)-driven sgRNA and HDR template were subcloned between inverse terminal repeat (ITR) sequences. (G) Neonatal cardiomyocytes isolated from Cas9 knock-in mice were transduced with AAV6 encoding editing components shown in (F) (1.88 × 105 viral genomes/cell). Forty-eight hours after transfection, genomic DNA was extracted, and the targeted region was amplified by PCR. Purified PCR products were digested with XhoI. Arrowheads indicate the cleaved PCR products. NTD: non-transduced control. (H) PCR products shown in (G) were cloned and sequenced. Proportion of HDR, NHEJ (cleavage sites ranging or not ranging to exon 13), and intact sequence were evaluated (50 clones each, n = 4, means ± SD).