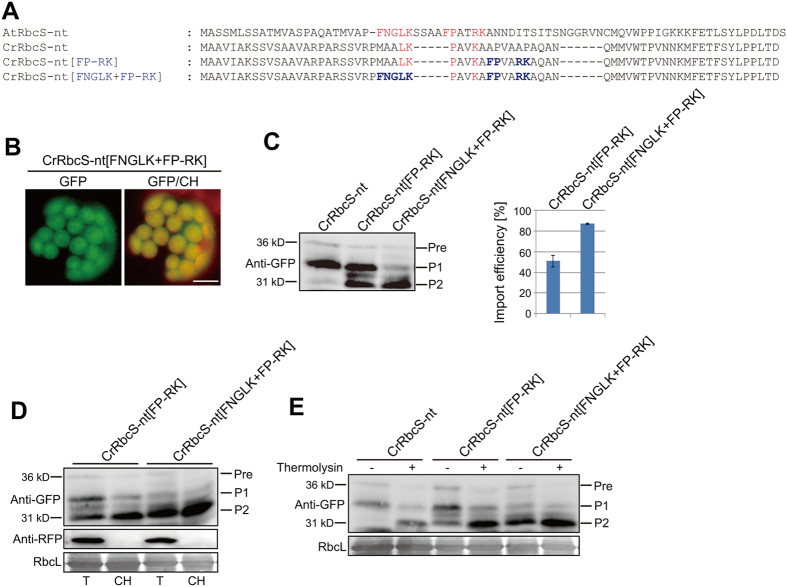
Figure 5.
Two sequence motifs, FNGLK and FR-PK, of AtRbcS TP further improve the ability of CrRbcS-nt to import proteins into plant chloroplasts. (A) Sequences of AtRbcS TP, CrRbcS TP, and modified CrRbcS TPs. All constructs were fused to GFP. (B) Localization of reporter proteins. Protoplasts from Arabidopsis plants were transformed with the indicated constructs, and GFP patterns were observed 12 h after transformation. Green, red, and yellow signals represent GFP, chlorophyll autofluorescence, and the overlap between green and red signals, respectively. Scale bar = 20 μm. (C) Western analysis of the reporter proteins. Total protein extracts from transformed protoplasts were analyzed by western blotting using anti-GFP antibody. Pre, precursor form; P1, processed form 1; P2, processed form 2. Signal intensity of protein bands was measured using LAS3000 imager (FUJI FILM) software, and import efficiency was defined as described in Fig. 3G. Error bar = SD (n = 3). (D) Isolation of chloroplasts from transformed protoplasts. Chloroplasts were isolated from the protoplasts transformed with the indicated constructs. At 12 h after transformation, protoplasts were gently lysed and chloroplasts were isolated using a Percoll gradient. Total and chloroplast fractions were analyzed by western blotting using anti-GFP and anti-RFP antibodies. RFP was used as a control for cytosolic proteins. Rubisco complex large subunit (RbcL) stained with Coomassie brilliant blue was used as a loading control. T, total protein; CH, chloroplast fraction; Pre, precursor form; P1, processed form 1; P2, processed form 2. (E) Thermolysin sensitivity of reporter proteins. Protoplasts transformed with the indicated constructs were gently lysed and treated with thermolysin. Protein extracts were analyzed by western blotting using anti-GFP antibody. Pre, precursor form; P1, processed form 1; P2, processed form 2.