Figure 4.
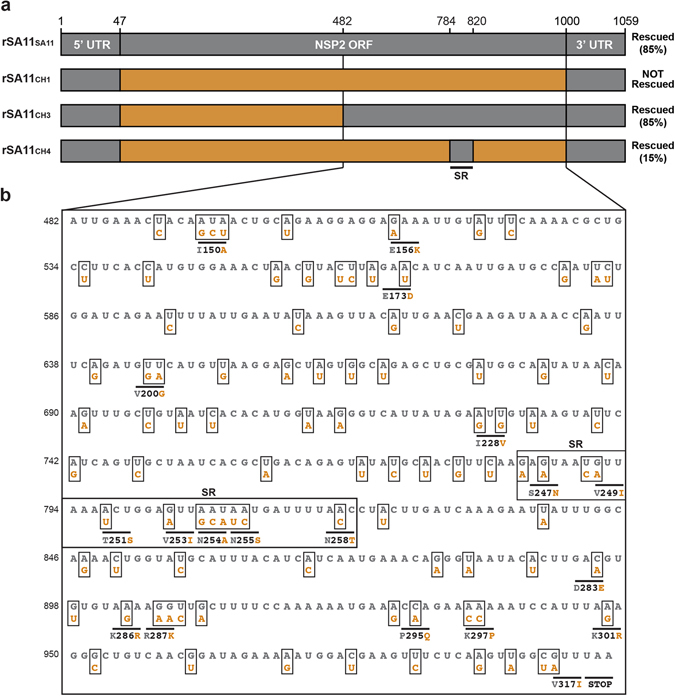
Nucleotide and amino acid differences between SA11 NSP2 and AU-1 NSP2 in the 482–1000 region. (a) NSP2 genes of recombinant reassortant viruses with wildtype SA11 or chimeric NSP2 genes are shown as boxes (not drawn to scale). For all genes, SA11 sequence is shown in grey and AU-1 sequence is shown in orange. Nucleotide numbers are listed at the top and the general locations of untranslated regions (UTRs) and open-reading frame (ORF) in the NSP2 gene are shown. The results of the rescue experiments, including % recombinant plaques, are listed to the right of each gene. (b) The sequence of nucleotides 482–1000 of SA11 NSP2 are shown in grey. The nucleotides that differ for AU-1 NSP2 are shown below the SA11 sequence and are colored orange. Amino acid changes are listed beneath the corresponding codon with the SA11 residue shown in grey and the AU-1 residue shown in orange. The SR (nucleotides 784–820) is outlined with a box and labeled.
