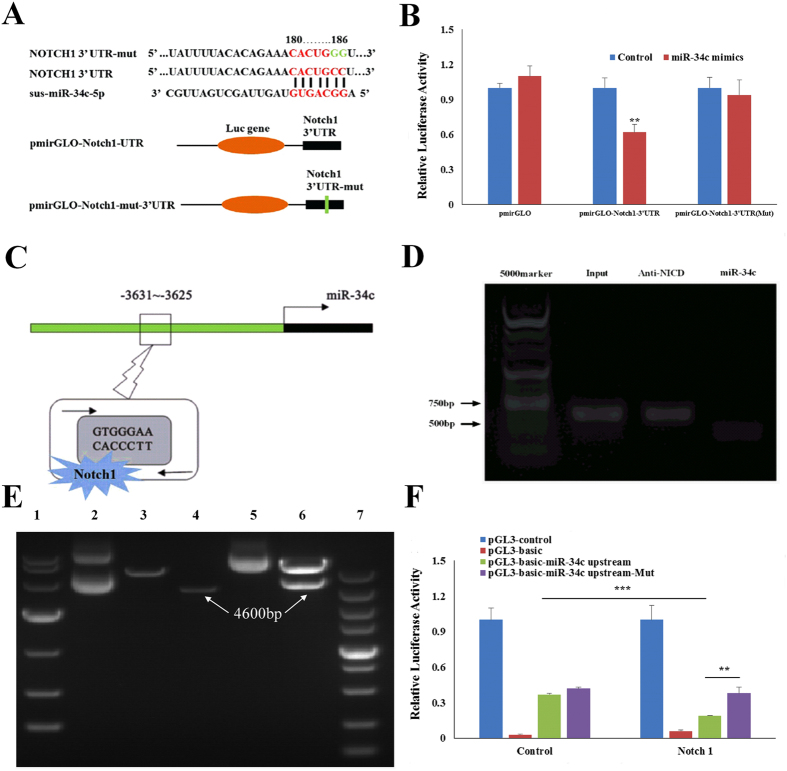
Figure 6.
Negative feedback between miR-34c and Notch1. (A) Schematic representation of the sequence of Notch1 3′UTR, Notch1 3′UTR-mut, miR-34c, and luciferase reporter constructs. (B) Luciferase reporters shown in (A) were transfected into HEK-293T cells with either miR-34c mimics or Control respectively. Luciferase activity was determined 24 h after transfection. (C) Schematic representation of Notch1 binds at the upstream of miR-34c in the genome. (D) PSCs transfected with pEGFP-N1ICD and were cultured in growth medium for 24 h. These PSCs were immune precipitated with the Notch1 antibody. The precipitated DNA was used to amplify the fragments of -3631 upstream of miR-34c gene by primers (arrows). (E) Cloned miR-34 upstream of its genomic site (about 4600 bp, the fourth lane), and constructed pGL3-basic-miR-34 upstream recombinant vector (the fifth and sixth lanes). Lanes 1–7 represent DL 10000, pGL3-basic, double digested pGL3-basic, double digested miR-34 upstream of its genomic site, pGL3-basic-miR-34 upstream recombinant vector, double digested pGL3-basic-miR-34 upstream recombinant vector, DL5000, respectively. (F) N1ICD decreased the pGL3-basic-miR-34 upstream recombinant vector relative luciferase activity. N1ICD with pGL3-basic-miR-34 upstream recombinant vector relative or pGL3-basic-miR-34 upstream (mut) recombinant vector were transfected into HEK-293T cells respectively. Luciferase activity was determined 24 h after transfection. *p < 0.05; **p < 0.01; ***p < 0.001; Student’s t-test. The results are presented as Mean ± S.E.M. of three replicates for each group.