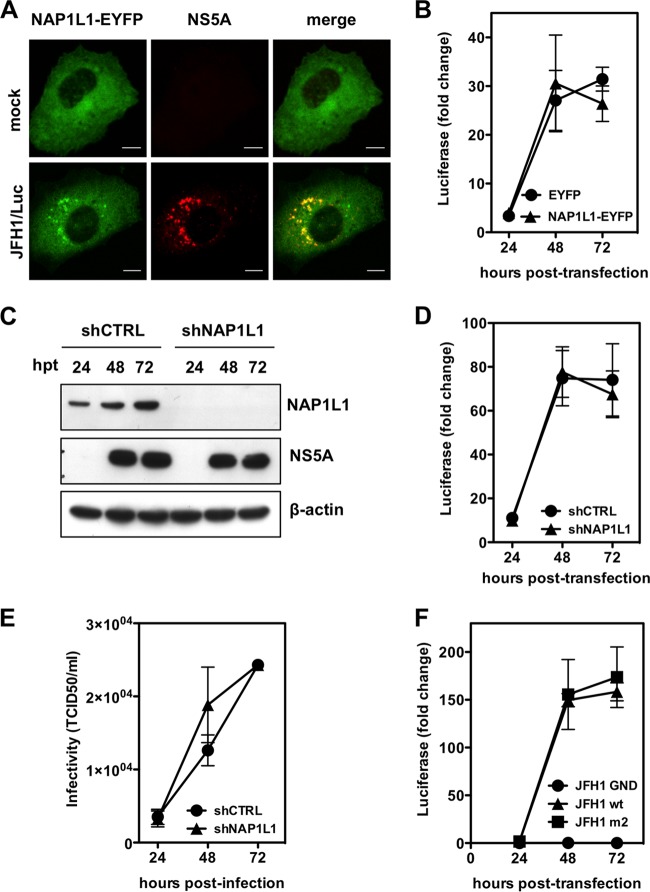
FIG 4.
HCV does not require NAP1L1 for replication and infectivity. (A) NS5A and NAP1L1-EYFP colocalize during HCV replication. Huh7-Lunet cells were transduced with a lentiviral vector (LV) expressing NAP1L1-EYFP and then either mock electroporated or treated with the subgenomic HCV replicon SGR-JFH1/Luc and fixed at 72 hpe. Indirect immunofluorescence analysis was performed with anti-NS5A antibody (red) (scale bars, 10 μm). (B) NAP1L1 overexpression does not affect HCV genome replication. Huh7-Lunet cells were transduced with a lentiviral vector (LV) expressing EYFP or NAP1L1-EYFP at an efficiency of >85% on average as measured by cytofluorimetric analysis. Cells were then electroporated with the HCV SGR JFH1/Luc RNA, and luciferase was monitored at the indicated time points. Values are normalized to the luciferase signal at 4 h postelectroporation. Averages for 3 independent replicates are shown with standard deviations. (C) Depletion of NAP1L1 by shRNA. Huh7-Lunet cells were transduced with an LV expressing shRNA targeting NAP1L1 (shNAP1L1) or a nontargeting control (shCTRL). After selection with puromycin, cells were electroporated with HCV SGR JFH1/Luc RNA, and protein levels were detected by WB. (D) Depletion of NAP1L1 does not affect HCV genome replication. Huh7-Lunet cells were treated as for panel C, and the luciferase signal was measured as for panel B. (E) Depletion of NAP1L1 does not affect HCV infectivity. Huh7-Lunet cells treated with shRNAs as for panel C were electroporated with full-length HCV JFH1 RNA. At the indicated time points, the supernatant was collected and used to infect naive Huh7.5 cells at various dilutions. The 50% tissue culture infective dose (TCID50) was then calculated by counting cells stained with the NS5A antiserum. (F) Mutagenesis of the NAP1L1 binding site of NS5A does not affect HCV genome replication. Huh7-Lunet cells were electroporated with HCV SGR JFH1/Luc RNA from the wild type (wt), the m2 mutant of NS5A, or the GND mutant of NS5B, which is replication defective. Luciferase was measured as for panel B.