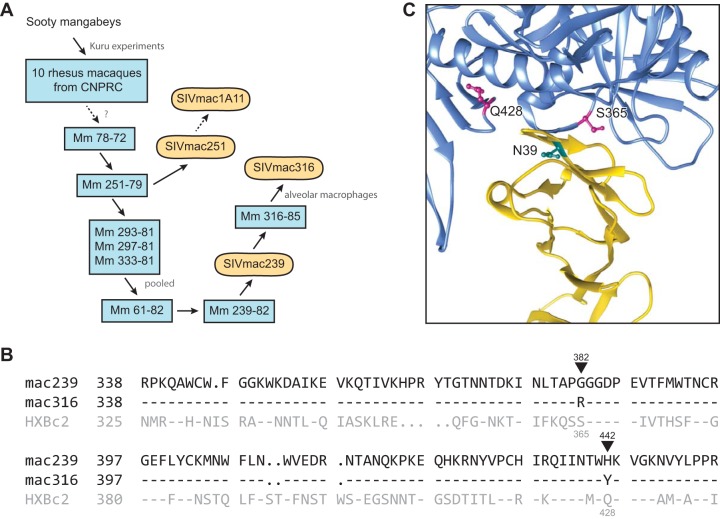
FIG 2.
Origins of and CD4-binding site differences between SIVmac239 and SIVmac316. (A) Derivation of some commonly studied SIVmac isolates. Blue rectangles represent rhesus macaques in which virus derived from sooty mangabey was passaged. Rounded rectangles represent uncloned biological isolates or infectious molecular clones. Solid arrows indicate direct isolation steps or viral transmission, and dotted arrows represent passaging and/or cloning steps omitted for simplicity. CNPRC, California National Primate Research Center (1, 44). (B) Sequence alignment of regions of the SIVmac239, SIVmac316, and HXBc2 Env protein participating in CD4 binding. Residues conserved with SIVmac239 are indicated by dashes. Black triangles indicate the two differences between SIVmac239 and SIVmac316 Envs in their CD4-binding sites. SIVmac239 numbering (black) and HXBc2 numbering (gray) is also provided. (C) The crystal structure (PDB accession no. 1G9M [45]) of CD4 (yellow) in complex with HXBc2 gp120 (blue) is shown. HXBc2 CD4-binding site residues S365 and Q428, corresponding to SIV residues 382 and 442, respectively, are indicated (magenta). The location of N39 of human CD4 is also indicated (cyan). Note the proximity of CD4 residue 39 to the CD4-binding site differences between SIVmac239 and SIVmac316.