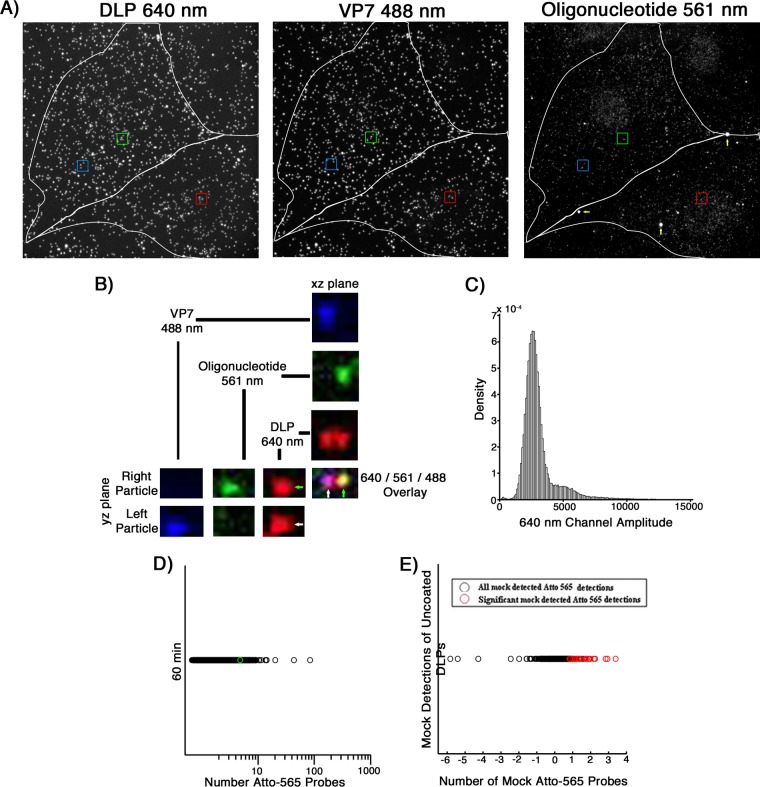
FIG 9.
In situ hybridization after 60 min of infection of MA104 cells at an MOI of 20. (A) Cells were infected at an MOI of 20 for 60 min with doubly labeled rcTLPs. Paraformaldehyde fixation was followed by overnight incubation with the pool of 44 Atto 565-labeled oligonucleotide probes and subsequent 3D imaging at 640-nm (left), 488-nm (middle), and 561-nm (right) excitation wavelengths, representative maximum-intensity projections of which are shown. Red, green, and blue boxes, DLPs colocalized with ∼4 Atto 565-labeled oligonucleotides. White arrows, likely oligonucleotide aggregates. (B) Fluorescently labeled particles in the green box in panel A, displaying the individual channels in the xz and yz planes passing through the center of each particle, as well as the maximum projection of the overlay of all three channels. Green arrow, an uncoated DLP that colocalizes with ∼4 Atto 565-labeled oligonucleotides. White arrow, an intact fluorescently labeled rcTLP with no colocalized oligonucleotide signal. (C) Probability density of 640-nm channel amplitudes derived from image analysis as described in Materials and Methods. Detections with amplitudes of less than 900 were not considered DLPs; detections above 4,000 were considered aggregates or multiple unresolvable DLPs. (D) Scatter plot of the number of Atto 565 probes colocalized with a given uncoated DLP after 60 min of infection at an MOI of 20. The particle highlighted in the green box in panel A and shown in B is represented here as a green circle plotted within the data. (E) In images collected from mock-infected cells probed with the Atto 565-labeled oligonucleotide pool, 30 random “DLP” locations were generated, matching the average number of uncoated DLPs calculated from the data in Table 3. Of the 2,460 mock particles (black circles), 28 (red circles) colocalized with significant signal in the 560-nm channel as described in Materials and Methods. Quantification of the number of colocalized Atto 565-labeled probes was performed as described in Materials and Methods.