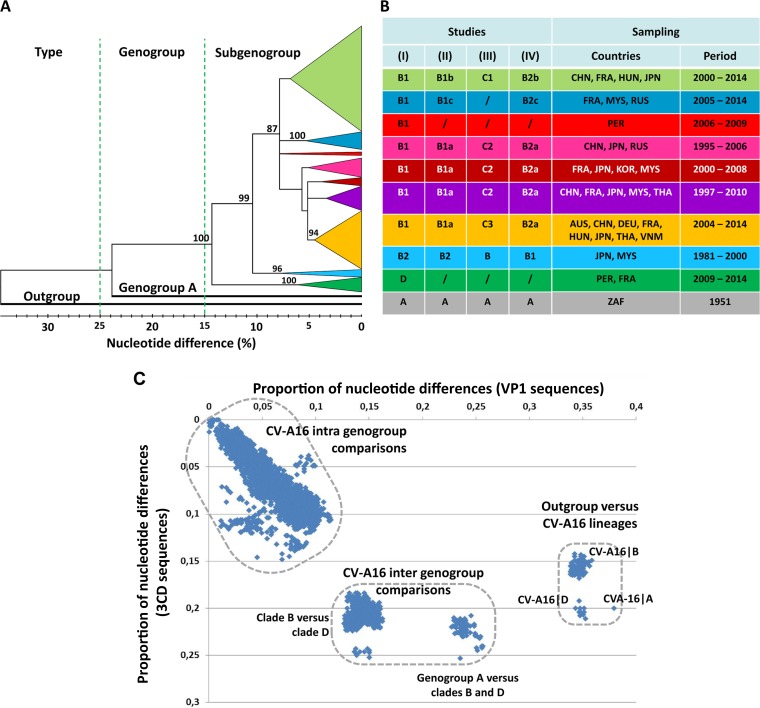
FIG 2.
CV-A16 genetic diversity assessed with the VP1 gene sequences. (A) Phylogenetic tree based on complete VP1 coding sequences of 573 CV-A16 sequences. The tree was inferred with the neighbor-joining method from genetic distances calculated with the p-distance algorithm. The tree topology was assessed with 1,000 bootstrap pseudoreplicates. The bootstrap values are shown only for the main phylogenetic clusters. The branches of consistent phylogenetic clusters were compressed for more clarity. The clade(s) (genogroup[s]), geographic origin(s), and isolation date(s) of virus strains are indicated for each cluster. The CV-A16 strain G10 (South Africa, 1951) is the only sample assigned to clade A (CV-A16∣A). The EV-A71 BrCr sequence was used as an outgroup. Distance thresholds used for demarcation of phylogenetic clusters are indicated with dashed lines. (B) Variations in the designations of CV-A16 phylogenetic groups (genogroups and subgenogroups) in the literature. (Studies I to IV) The table shows the correspondence between phylogenetic data reported in the present study (I) and the CV-A16 groups described in the following other studies: Zhang et al. (27) and Sun et al. (28) (II), Li et al. (31) and Iwai et al. (29) (III), and Zong et al. (30) (IV). A number of discrepancies in the designations of the main CV-A16 phylogenetic groups are indicated./,not applicable; AUS, Australia; CHN, China; DEU, Germany; FRA, France; HUN, Hungary; JPN, Japan; KOR, South Korea; MYS, Malaysia; PER, Peru; ZAF, South Africa; RUS, Russia; THA, Thailand; VNM, Vietnam. (C) Scatterplot comparing the pairwise nucleotide distances calculated with the VP1 and 3CD gene sequences of a sample of 181 CV-A16. The sample comprised 111 strains of the present study and 70 strains for which sequence data are publicly available. The main genetic clusters are delimited by dashed lines.