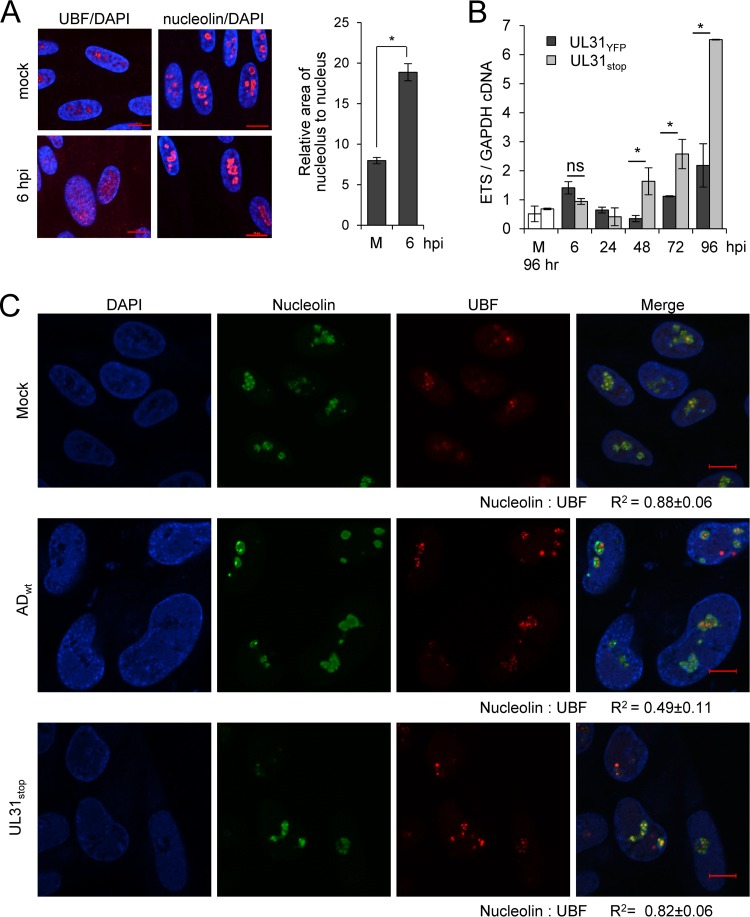
FIG 6.
CMV pUL31 alters nucleolar organization and function during infection. (A) MRC-5 fibroblasts were mock treated or infected at an MOI of 3, fixed at 6 hpi, and stained using antibodies to UBF (red; left) or nucleolin (red; right) and DAPI (blue). Data represent the average change in area of nucleoli relative to nucleus from mock-infected (m) or infected cells at 6 hpi. Nucleolin and DAPI staining were used to define perimeters of nucleoli and the nucleus, respectively. The relative area of nucleolus to nucleus per cell (in pixels) was determined and averaged from 50 cells. (B) Fibroblasts were infected with UL31YFP or UL31stop virus or mock infected at an MOI of 3, and total RNA was isolated at the indicated times. Mock-infected samples were collected at 96 h. Pre-rRNA levels were determined using qRT-PCR and primers against the 5′ external transcribed spacer (ETS) of rRNA relative to GAPDH. Data are from three biological and two technical replicates, with error bars representing standard deviations from the means. (C) Fibroblasts were mock treated or infected with the parental UL31YFP or UL31stop virus at an MOI of 3. At 96 hpi, samples were fixed and stained with antibodies to nucleolin or UBF. Colocalization was calculated by defining a region of interest determined by nucleolin staining from 15 to 20 cells, and the average Pearson correlation coefficient (R2) between the indicated signals was determined.