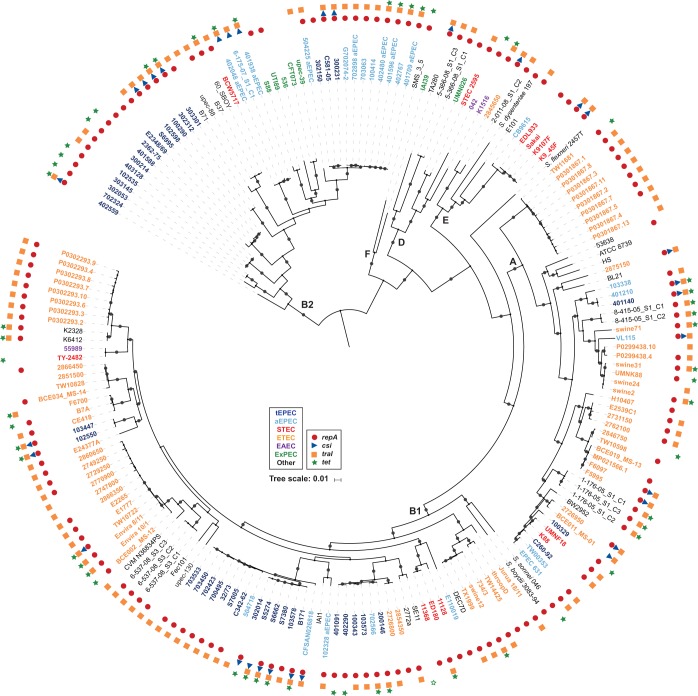
FIG 2.
Phylogenomic analysis of E. coli genomes that carry genes with similarity to those of pB171_90. A SNP-based phylogeny was generated using ISG as previously described (35, 70). There were 149,150 conserved SNP sites identified among all of the E. coli genomes analyzed relative to the reference E. coli strain IAI39 (GenBank accession number NC_011750.1) that were used to construct a maximum-likelihood phylogeny with 100 bootstrap values using RAxML v.7.2.8 (71). The phylogeny was midpoint rooted and labeled using iTOL v.3 (72). Bootstrap values of ≥80 are designated by a gray circle. The scale bar indicates the approximate distance of 0.01 nucleotide substitutions per site. The E. coli phylogroups B1, A, B2, D, E, and F are denoted by bold letters. Symbols designate the presence of pB171_90 genes in each genome with tetA and tetR both represented by green stars (both genes, solid green star; one gene, open green star). Colors of the genome labels indicate the presumptive pathotype that was determined by in silico detection of canonical virulence genes.