Figure 2.
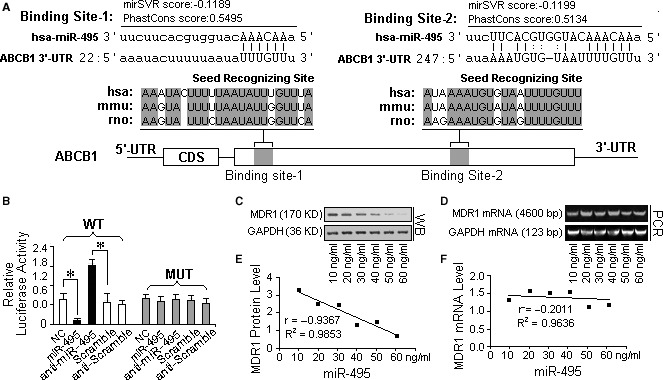
ABCB1 was identified as a direct target of miR‐495. (A) A schematic description of the hypothetical duplexes formed by the interactions between the binding sites in the ABCB1 3′‐UTR and miR‐495. The mirSVR scores (−0.1199, −0.1199) and PhastCons scores (0.5495, 0.5134) of the two hybrids are within the range of genuine miRNA‐target pairs. Two seed recognition sites were found in the 3′‐UTR, and the nucleotides in these regions are highly conserved across humans, mice and rabbits. (B) The luciferase reporter activity of the vector containing the mutated miR‐495 binding sites in the ABCB1 3′‐UTR was unaffected by miR‐495. In contrast, the luciferase reporter activity of the plasmid containing the wild‐type MDR1 3′UTR sequence was increased more than 75% in A2780DX5 cells cotransfected with a transfection control plasmid (β‐gal) and anti‐miR‐495, but it was unaffected by the knockdown of miR‐495, compared with the cells treated with the negative control RNA, suggesting a specific binding between miR‐495 and the mRNA of MDR1. (C) Dose‐dependent changes in the expression of the MDR1 protein in A2780DX5 cells expressing the miR‐495 mimic. (D) Dose‐dependent changes in the expression of the MDR1 mRNA in A2780DX5 cells transfected with the miR‐495 mimic. (E and F) Pearson's correlation scatter plots of the fold change of the levels of miR‐495 and ABCB1 protein or mRNA in A2780DX5 cells. There is an inverse correlation between the miR‐495 levels and MDR1 levels, but no significant difference can be observed between the MDR1 mRNA levels of the differently treated cells, implying that miR‐495 inhibited the translation of the MDR1 mRNA but that it did not induce degradation of the mRNA itself. *Student's t‐test (paired, one tailed), P < 0.05, n = 3.
