Fig. 1.
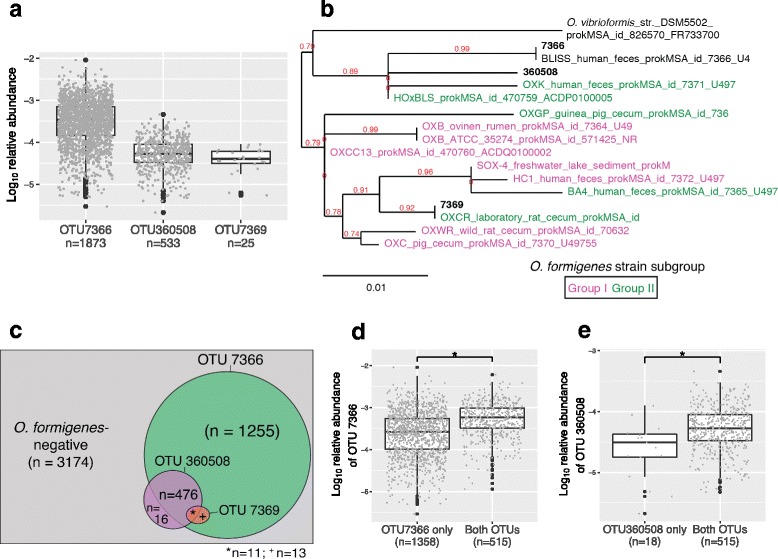
Co-occurrence and abundances of three O. formigenes-associated OTUs in fecal samples from 4945 subjects. a Abundance of three O. formigenes-associated OTUs (7366, 360508, 7369) in samples with colonization detected, by closed-reference OTU picking. b O. formigenes phylogenetic tree. The tree was built from 16S V4 region sequences of three O. formigenes-associated OTUs (bold): 13 O. formigenes strains with group I (purple), group II (green), and strain BLISS with group unknown (black); O. vicrioformis selected from the Oxalobacter family used as an outgroup. The tree was built based on maximum likelihood with log-likelihood of −1317.6. Branch support values designated in red. Statistical details of the tree are included in the Additional files 9 and 10. c Euler diagram of co-occurrences in the 4945 subjects. The 184 subjects with multiple samples were considered positive if any sample was positive for at least one O. formigenes OTU. d, e Abundance of the studied OTUs in the samples in which either one or both OTUs were present. Panels focus on OTU 7366 (d) or 360508 (e). One-sided Mann–Whitney tests were used to determine whether or not the indicated OTU is more abundant when both are present, *p < 0.001, by Mann–Whitney test
