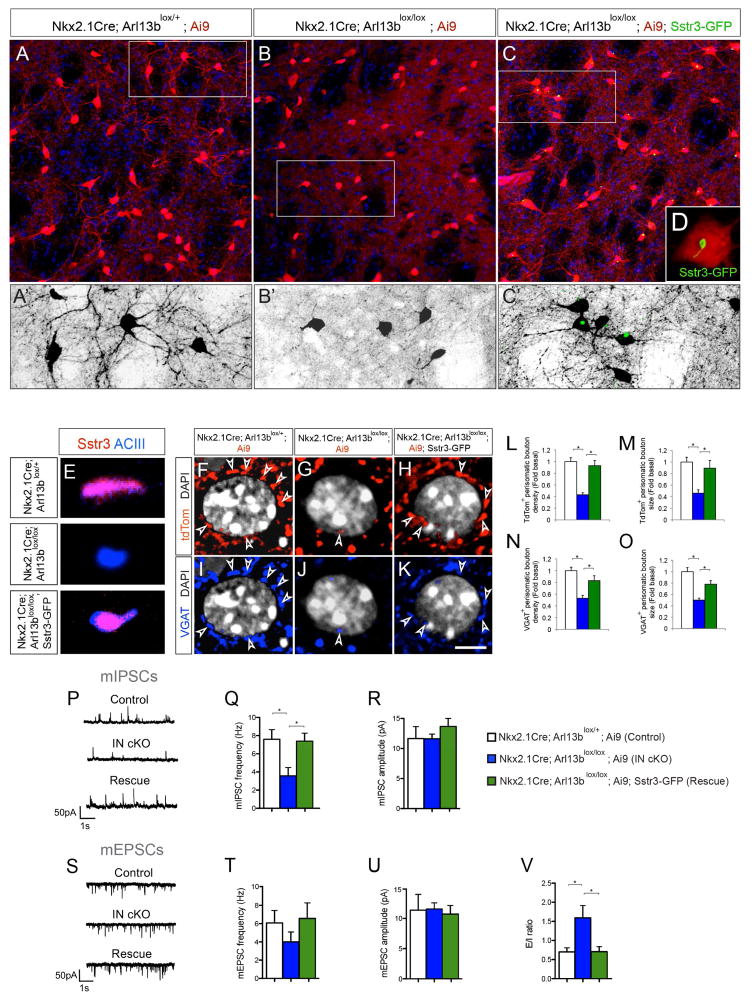
Figure 5. Primary cilia-driven Sstr3 signaling regulates interneuron morphology and connectivity.
(A–C) Striatal tdTom+ INs in Nkx2.1Cre; Arl13blox/+; Ai9 (A), Nkx2.1Cre; Arl13blox/lox; Ai9 (B), and Nkx2.1Cre; Arl13blox/lox; Ai9; Sstr3-GFP (C) brains [P30]. (A′–C′) Higher-magnification images of tdTom+ striatal INs from outlined area in panels A–C. (D) Induced Sstr3-GFP expression in primary cilia of Nkx2.1Cre; Arl13blox/lox; Sstr3-GFP; Ai9 INs. (E) Altered Sstr3 localization in Arl13b deficient IN primary cilia and re-expression after induction of Sstr3-GFP. Co-labeling of primary cilia with anti-ACIII and anti-Sstr3 antibodies in Nkx2.1Cre; Arl13blox/+; Ai9, Nkx2.1Cre; Arl13blox/lox; Ai9 and Nkx2.1Cre; Arl13blox/lox; Ai9+; Sstr3-GFP INs. (F–K) Perisomatic boutons (arrowheads) labeled with tdTom (F–H) and anti-VGAT antibodies (I–K) in Nkx2.1Cre; Arl13blox/+; Ai9 (F, I), Nkx2.1Cre; Arl13blox/lox; Ai9 (G, J) and Nkx2.1Cre; Arl13blox/lox; Sstr3-GFP; Ai9 (H, K) brains [P30]. (L–M) Quantification of tdTom+ perisomatic bouton density (L) and size (M). Data shown are mean ± SEM. *P<0.05 (One-way ANOVA: F2,42 [L] = 23.0; p = 5.53616E-07; post-hoc p[L, lox/+ vs. lox/lox] = 6.08809E-08, post-hoc p[L, lox/lox vs. lox/lox-Sstr3] = 3.26839E-05; F2,42 [M] = 6.54; p = 0.004; post-hoc p[M, lox/+ vs. lox/lox] = 3.64284E-05, post-hoc p[M, lox/lox vs. lox/lox-Sstr3] = 0.008). n = 15 cells from 3 different brains per group. (N–O) Quantification of VGAT+ perisomatic bouton density (N) and size (O). Data shown are mean ± SEM. *P<0.05 (One-way ANOVA: F2,42 [N] = 13.35; p = 5.6345E-05; post-hoc p[N, lox/+ vs. lox/lox] = 5.21332E-06, post-hoc p[N, lox/lox vs. lox/lox-Sstr3] = 0.004; F2,42 [O] = 17.68; p = 6.04031E-06; post-hoc p[O, lox/+ vs. lox/lox] = 4.23172E-06, post-hoc p[O, lox/lox vs. lox/lox-Sstr3] = 0.0009). n = 15 cells from 3 different brains per group. (P–U) Patch-clamp electrophysiological recordings of striatal MSNs from Nkx2.1Cre; Arl13blox/+; Ai9 (control, n=6), Nkx2.1Cre; Arl13blox/lox; Ai9 (mutant, n=10) and Nkx2.1Cre; Arl13blox/lox; Sstr3-GFP; Ai9 (rescue, n=4) brains. (P, S) Representative patch-clamp electrophysiological recordings from MSNs showing mIPSCs (P) and mEPSCs (S). (Q, R) Quantification of mIPSC frequency (Q) and amplitude (R). *P<0.05 (One-way ANOVA, mIPSC frequency: F2,20 = 7.7; p = 0.003; post-hoc p[ctrl vs. IN cKO] < 0.01, post-hoc p[Rescue vs. IN cKO] < 0.05; control, n = 6; IN cKO, n = 11; rescue, n = 6). (T, U) Quantification of mEPSC frequency (T) and amplitude (U). (V) Rescue of overall excitatory/inhibitory ratio in striatal MSNs of Nkx2.1Cre; Arl13blox/lox; Sstr3-GFP; Ai9 mice. Data shown are mean ± SEM. *P<0.05 (One-way ANOVA: F2,19 = 4.8; p = 0.021; post-hoc p[ctrl vs. IN cKO] < 0.05, p[Rescue vs. IN cKO] < 0.05; control, n = 6; IN cKO, n = 10; rescue, n = 6). Scale bar, 37.6μm (A–C); 18μm (A′–C′); 6.2μm (D); 1.3μm (E); 5μm (F–K). See also Figure S5.