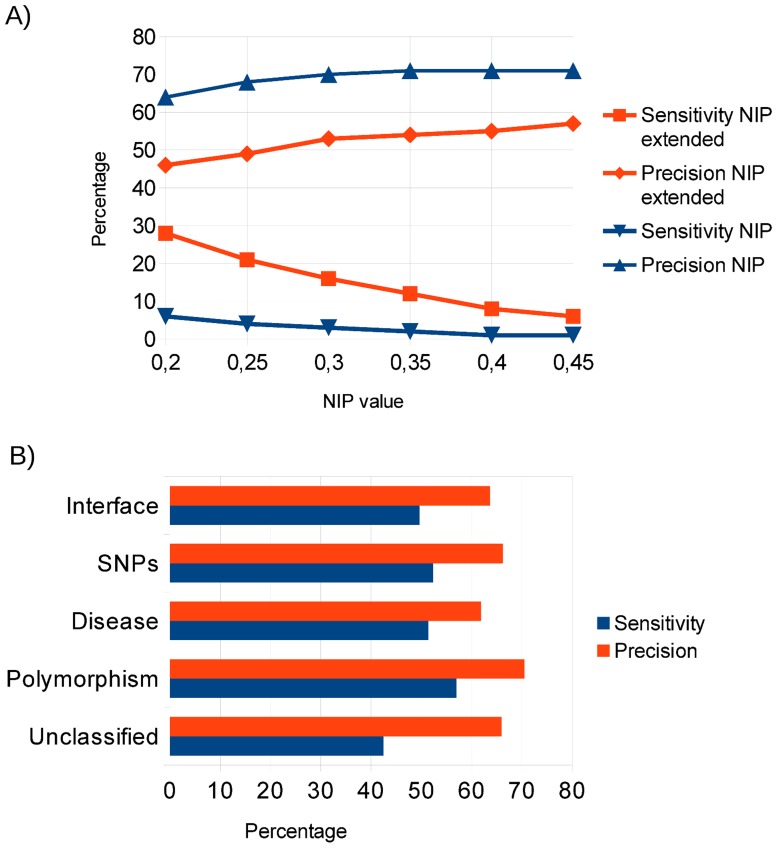
Fig 2. Prediction of interface residues and nsSNPs.
(A) Prediction success (sensitivity and precision) of interface residues using pyDockNIP (alone or extended with neighbor residues) on the proteins of the protein-protein docking benchmark 4.0, according to NIP cutoff value. (B) Interface and nsSNPs predictions using the extended pyDockNIP predictions on the proteins of the structural interaction networks from the six selected diseases. The nsSNPs predictions are detailed for interface disease-related, polymorphism and unclassified nsSNPs.