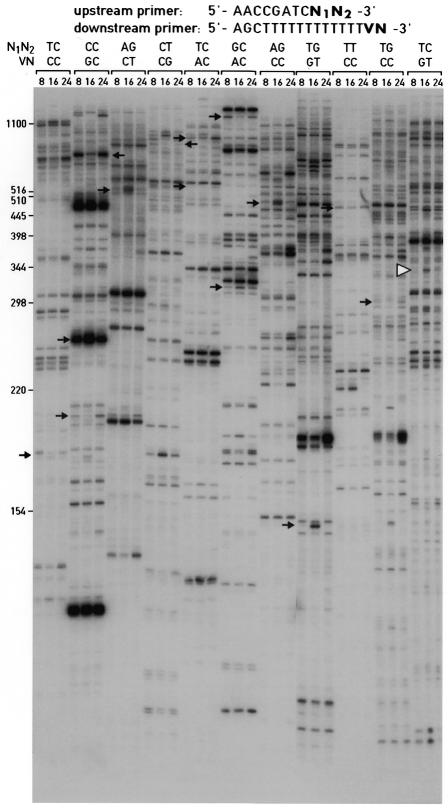
Figure 3.
Differential display of 3′-terminal ds cDNA fragments. Aliquots of ds cDNA libraries were amplified with the display primers shown and size-fractionated on a 5% urea–polyacrylamide gel alongside size markers (left hand side). The times at which the animals were sacrificed to prepare liver RNA are given at the top. The autoradiography was exposed for 72 h at room temperature without an intensifier screen. The positions of cDNA fragments that were cloned and sequenced are indicated by arrows. All of the mRNAs complementary to these cDNAs showed circadian accumulation profiles, when examined by northern blot hybridization or ribonuclease protection assays (Fig. 5 and data not shown). One of the cloned cDNA fragments, GNCLP (indicated by a white arrowhead), was used to examine the sensitivity and priming specificity of the ADDER method (Figs 5 and 6). Several additional fragments for potentially circadian mRNAs can be seen in the autoradiograph; however, these fragments have not yet been cloned and the corresponding mRNAs have thus not been examined for cyclic accumulation by northern blot or RNase protection assays.