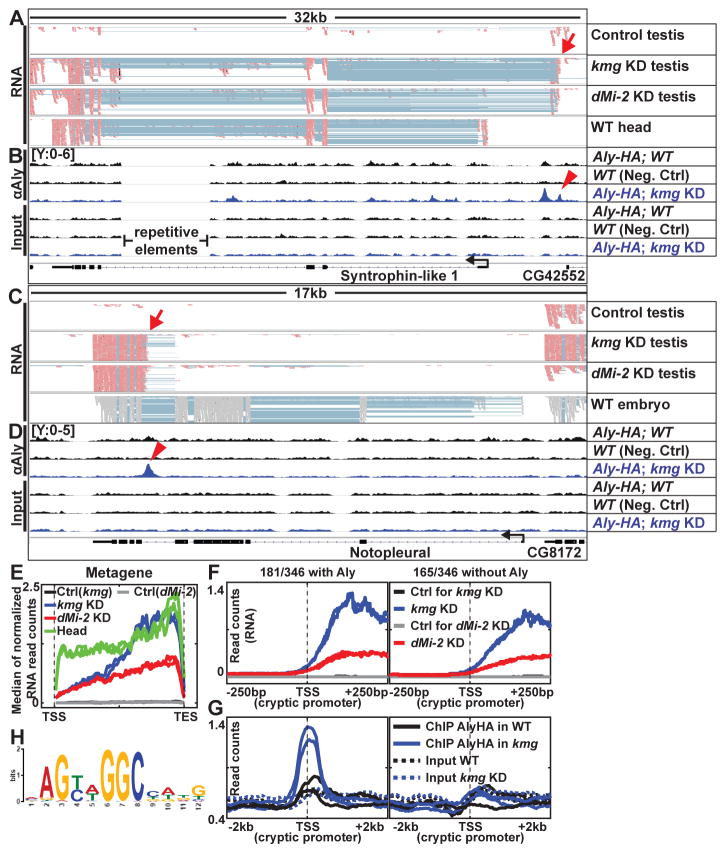
Figure 4. Kmg and dMi-2 prevent misexpression from cryptic promoters.
(A and C) Genome browser screenshots of RNA-Seq reads mapped to Crick (−) strand (pastel red) or mapped without strand information (gray) (embryo data from modENCODE) (27). Pastel blue lines: a single read that skipped an intron and mapped to two adjacent exons. Red arrows: cryptic promoter sites.
(B and D) Aly-HA ChIP and input read density in wild-type (with and without Aly-HA transgene) and kmg KD testes for the same regions. Red arrowheads: peak of enrichment by ChIP for Aly coinciding with potential cryptic promoter sites.
(E) Median RNA expression profile of 143 genes expressed in wild-type heads but not in wild-type testes and misexpressed in kmg KD testes, plotted as a metagene excluding introns. TSS: Transcription start site. TES: Transcription end site.
(F and G) Median profile of (F) RNA-Seq and (G) Aly ChIP (solid lines) and corresponding input (dotted lines) signals centered at cryptic promoters.
(H) Position weight matrix representation of sequence motif most significantly enriched in a collection of 181 cryptic promoters (±150bp around the TSS) with peaks of Aly binding in kmg KD testes.