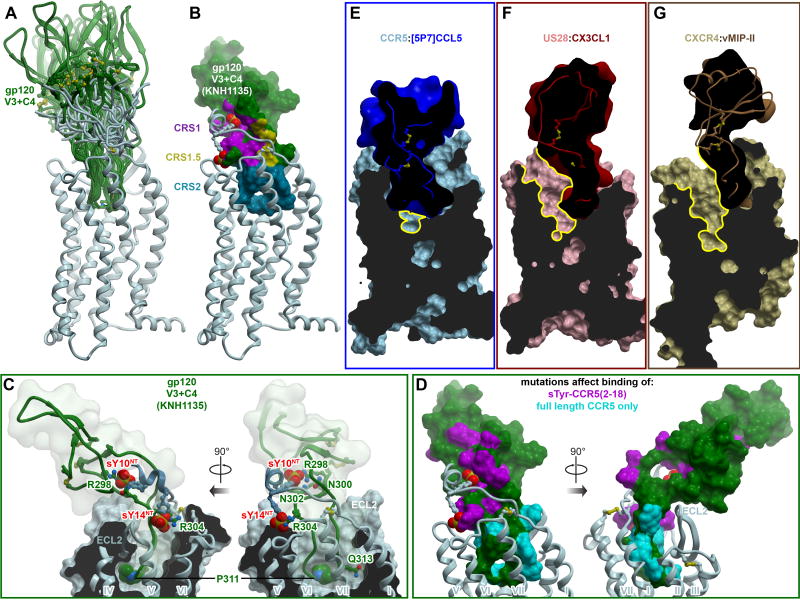
Figure 7. Insights into CCR5 interaction with HIV gp120 and inhibition of this interaction by chemokines.
(A) Ensemble of models of complexes between CCR5 and a fragment of HIV gp120 containing its V3 loop and C4 region. The ensemble illustrates multiple possible orientations of gp120 with respect to the receptor, enabled via the inherent flexibility of the interaction interface.
(B) Representative model of CCR5 (ribbon) in complex with the V3/C4 fragment of HIV gp120 (strain KNH1135, molecular surface). Interaction sites for the receptor N-terminus, the conserved 19-PC-20 motif, and the TM binding pocket are highlighted to illustrate architectural similarity to complexes with chemokines.
(C) Same model as in (B), showing interactions of important residues. CCR5 is shown in ribbon and a cut-away molecular surface, gp120 in ribbon surrounded by a transparent simplified molecular surface. Residues mentioned in the text are shown in sticks. Residue P311 in the conserved GPG[RQ] motif of the gp120 V3 crown, as well as the CCR5 N-terminal tyrosine sulfates are shown as spheres.
(D) Prior mutagenesis of gp120 mapped onto the surface of the gp120 V3/C4 fragment using the same model as in (B). Residues that affect binding of a sulfo-tyrosinated CCR5 N-terminal peptide (2–18) are colored in magenta. Residues whose mutations show effects only in the context of full-length CCR5 are show in cyan. The predicted architecture and topology of the complex are highly consistent with mutagenesis.
(E-F) Among chemokine crystallized with receptors, [5P7]CCL5 is unique in its ability to entirely fill the binding pocket of CCR5 (E). By contrast, binding of CX3CL1 to US28 (F) or vMIPII to CXCR4 (G) leads to only partial occupancy of the pocket, with much of the major subpocket remaining accessible (yellow contours). Receptors are shown as cut-away surfaces, chemokines as simplified cut-away surfaces and ribbons.
See also Figure S5, Table S5 and Table S6.