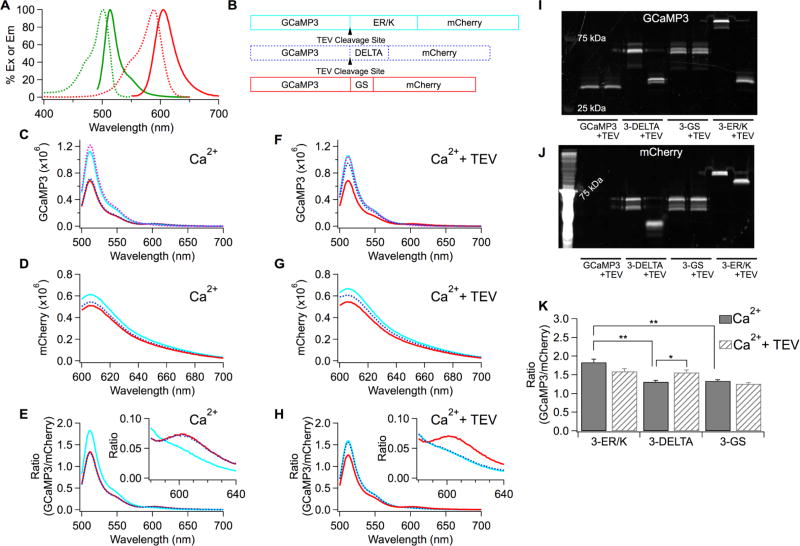
Figure 1.
Prevention of FRET between GCaMP3 and mCherry by ER/K helix. (A) Excitation (dash) and emission (solid) spectra of GCaMP3 (green) and mCherry (red) proteins. (B) Schematic drawing of tandem sensors with the different linkers. (C–H) Emission spectra of the tandem GCaMP3 and mCherry proteins in CaCl2 solution without TEV protease (C,D,E) or with TEV protease (F,G,H). Spectra of GCaMP3 alone (dashed pink) added to compare with those of the tandem sensors in panels C and F. (E,H) The peak intensity at 608 nm of the mCherry emission spectrum (D,G) was used to normalize the GCaMP3 emission spectrum denoted as Ratio (GCaMP3/mCherry) in each tandem sensor. Insets zoomed in on the spectra around 600 nm. (I,J) The protein samples used for obtaining the emission spectra (C–H) were run on a gel and visualized by exciting either GCaMP3 (I) or mCherry (J) to confirm the cleavage of GCaMP3 from mCherry. (K) Average peak ratios at 511 nm obtained from (E,H). Mean ± SEM (**p < 0.01, *p < 0.05, n = 3 for each condition).