Abstract
Pulex irritansL. is a cosmopolitan flea species that infests a wide variety of hosts. In North America it generally parasitizes large wild mammals, but in the Pacific Northwest an association has emerged between P. irritans and the western burrowing owl (Athene cunicularia hypugaea). While investigators have recognized this association for decades, it has not been clear if P. irritans feeds on burrowing owls, or if the owls serve exclusively as phoretic hosts. Here we describe using a real-time assay that was originally developed to identify bloodmeals in Ugandan cat fleas (Ctenocephalides felis Bouché) to detect burrowing owl DNA in P. irritans collected from burrowing owls in southern Idaho. Of 50 fleas tested, 12 had no detectable vertebrate bloodmeal. The remaining 38 (76%) contained burrowing owl DNA. The assay did not detect vertebrate DNA in unfed fleas exposed to owl or mouse pelts and is therefore unlikely to detect DNA in fleas from vertebrates that have served exclusively as phoretic hosts. We conclude that P. irritans feeds on burrowing owls. We discuss the potential implications of this finding for burrowing owl conservation and enzootic plague dynamics.
Keywords: Pulex irritans, burrowing owl, Athene cunicularia, bloodmeal identification, flea
Pulex irritans L., the so-called human flea, parasitizes a wide variety of hosts, including rodents, large wild mammals, and livestock (Gratz 1999). In North America it generally parasitizes large mammals, particularly carnivores, but specific host associations vary between geographic regions (Hopla 1980, Lewis et al. 1988). In the northwestern United States and southern British Columbia, investigators have observed a surprising association between P. irritans and western burrowing owls (Athene cunicularia hypugaea; hereafter burrowing owls), small, ground-dwelling owls of western North America (Hopla 1980, Smith and Belthoff 2001, Belthoff et al. 2015). Recently, Belthoff et al. (2015) reported that P. irritans comprised >99% of 4,791 fleas collected from breeding burrowing owls or their nestlings in study sites in Idaho, Oregon, and Washington. This phenomenon appears to occur primarily in the Pacific Northwest, although P. irritans have also been collected from burrowing owls in western Montana and northern Utah (Hopla 1980). Pulex irritans was the only species of flea collected from adult burrowing owls captured from one study site in Colorado, but only a single flea was collected from a single individual. No fleas were detected on 55 adult burrowing owls captured from another study site in South Dakota (Belthoff et al. 2015). Skoruppa et al. (2006) found no fleas on burrowing owls wintering in south Texas.
While investigations have shown a relationship between burrowing owls and P. irritans, it has not been clear if P. irritans feeds on the owls, or if the owls serve exclusively as phoretic hosts. Phoretic hosts provide transport for fleas but do not provide bloodmeals. The nature of this arthropod–host relationship could have implications for burrowing owl conservation. The United States Fish and Wildlife Service lists the burrowing owl as a national bird of conservation concern, and this species is endangered in Canada and threatened in Mexico (Klute et al. 2003). Ectoparasitism of bird species in decline merits concern because fleas can decrease nestling growth and survival, affect parental feeding rates and sleeping regimens, and prompt increased grooming and nest sanitation, which can take time and energy away from caring for young (Richner et al. 1993, Christe et al. 1996, Boughton et al. 2006, Cantarero et al. 2013). Hematophagous ectoparasites deplete nutrients which could otherwise contribute to host growth, maintenance, or reproduction. Moreover, flea saliva contains potent immunogens that induce energy-demanding immune responses in the host (Møller et al. 2005, Krasnov 2008). Thus, feeding fleas could challenge declining burrowing owl populations.
Whether or not P. irritans feed on burrowing owls may also have implications for plague ecology. P. irritans is known to be a competent vector of Yersinia pestis, the etiologic agent of plague (Verjbitski 1908, Blanc and Baltazard 1941), and it is incriminated as a Y. pestis vector in Madagascar, parts of Southern and Central Africa, South America, South Asia, and the Middle East (Gratz 1999, Drancourt et al. 2006, Laudisoit et al. 2007, Ratovonjato et al. 2014). While avian species are generally resistant to infection with Y. pestis (Meyer 1950), some investigators have suggested that birds of prey, including the burrowing owl, may play a role in sylvatic plague dynamics by transferring fleas between susceptible rodent populations (Jellison 1939, Wheeler et al. 1941, Brown 1944, Smith and Belthoff 2001). Previous work (Eisen et al. 2006, 2007) showed that taking an uninfected bloodmeal after taking an infectious one decreased the probability that ground squirrel fleas (Oropsylla montana Baker) would subsequently transmit Y. pestis to an uninfected host. This finding appears to be generalizable, at least within Oropsylla spp. (Wilder et al. 2008a, b). Therefore, taking an uninfected bloodmeal after taking an infectious one may have a similar effect on P. irritans transmission efficiency. If burrowing owls provide bloodmeals to P. irritans rather than serving exclusively as phoretic hosts, it may reduce the probability that a Y. pestis-infected P. irritans will subsequently transmit the bacterium to a susceptible host.
Graham et al. (2012, 2013) previously developed a real-time polymerase chain reaction (PCR)-based assay for identifying blood-meals in cat fleas (Ctenocephalides felis Bouché) collected from huts in the West Nile region of Uganda. Here we sought to determine if 1) This assay was flexible enough to identify burrowing owl blood, and 2) We could detect burrowing owl bloodmeals in P. irritans collected from burrowing owls in southern Idaho. To address these questions, we needed to ensure that if we did detect burrowing owl DNA in field-collected fleas it was not contaminating DNA from exposure to an owl’s skin and feathers. We therefore also sought to determine if our assay would detect vertebrate DNA in unfed fleas exposed to animal pelts.
Materials and Methods
Study Area, Owl Blood Collection, and Flea Collection and Identification
During May–June 2012, we collected fleas from burrowing owls (Fig. 1) nesting in artificial burrows within the Morley Nelson Snake River Birds of Prey National Conservation Area (NCA) in southwestern Idaho. This area is described in detail elsewhere (Smith and Belthoff 2001, Belthoff and King 2002). The population of burrowing owls in the NCA has been under long-term study, and fleas are prevalent on the owls (Smith and Belthoff 2001, Belthoff et al. 2015). As part of long-term monitoring, we also obtained ~ 100 μl of blood from owls using venipuncture of a wing vein. Blood was collected in micro hematocrit tubes, immediately transferred to 1.5-ml centrifuge tubes, stored on ice in the field, and then frozen at −20°C. The burrowing owl pelt we used (see below) was from a nestling that died in a nest burrow of unknown causes prior to fledging and was stored frozen at −20°C. Field research procedures for burrowing owls were approved by Boise State University’s Institutional Animal Care and Use Committee and authorized by appropriate federal and state permits issued to J. Belthoff.
Fig. 1.
A nesting pair of adult burrowing owls occupying an artificial burrow nest site within the Morley Nelson Snake River Birds of Prey National Conservation Area in southwestern Idaho. The tunnel entrance to the subterranean nest chamber is visible.
We used an aspirator and forceps to collect fleas from owls at one nest for the present study. We made no attempt to remove all fleas from the owls; rather, we sought to collect a sample. Fleas were held on ice in a plastic bag in the field and subsequently stored at −20°C. Fleas, pelts, and blood were shipped on dry ice to the Centers of Disease Control and Prevention Division of Vector-Borne Diseases (Fort Collins, CO) for analysis. We identified the fleas using morphologic keys (Furman and Catts 1982).
DNA Extraction, Real-Time PCR, and Sequencing
Only fleas with a visibly intact exoskeleton were used for bloodmeal analysis. We soaked each flea in 50% bleach (3.08% sodium hypochlorite) to remove surface contaminants, rinsed and homogenized it in calcium- and magnesium-free Dulbecco’s Phosphate Buffered Saline (Life Technologies, Grand Island, NY), extracted DNA as previously described (Graham et al. 2012), and eluted with 70 μl RT-PCR-grade water (Life Technologies). Each set of DNA extractions from field-collected fleas included a newly emerged (unfed) colony-reared Xenopsylla cheopis Rothschild (Division of Vector-Borne Infectious Diseases, Fort Collins, CO) as a negative extraction control. We stored all DNA at −80°C until PCR analysis. We extracted DNA from burrowing owl blood using the DNeasy Blood and Tissue Kit (Qiagen, Valencia, CA).
All templates were amplified and analyzed using a slightly modified version of a previously described SYBR Green I-based real-time PCR assay (Graham et al. 2012). Briefly, we employed M13-tagged primers that amplify an approximately 100-nt variable region of the 12S mitochondrial RNA gene in vertebrates but do not amplify flea DNA. Each reaction included the primers at a concentration of 135 nM each, 1X iQ SYBR Green Supermix (Bio-Rad Laboratories, Hercules, CA) and 10 μl template. A 5-min initial denaturation was followed by 40 amplification cycles. The final cycle was immediately followed by a melting analysis cycle. Each real-time run included at least one no template control (water) and positive control DNA isolated from Rattus norvegicus whole blood. The control DNA also served as an inter-run calibrator, ensuring equivalent threshold settings across runs. We ran all samples in duplicate. We purified and sequenced selected amplicons and generated a single sequence for each sample as previously described (Graham et al. 2012).
To verify that the assay detected burrowing owl DNA, we used DNA isolated from burrowing owl blood. By running 10-fold dilutions, five replicates per dilution, we determined that the assay amplified A. cunicularia hypugaea DNA and had a linear range of detection (LRD) of at least 1 pg–10 ng (R2 =0.999, efficiency =93.9%). The limit of detection (1 pg burrowing owl DNA) corresponded to a threshold cycle (Ct) value of 35.81. We selected the highest whole cycle value that fell within the LRD, 35, as our Ct cutoff for this study. We considered a sample positive if the collective Ct value (combined replicates) was ≤35, neither replicate had a Ct value >37, and both replicates yielded similar melting curves with single peaks. We repeated any sample that yielded dissimilar replicates or melting curves with multiple peaks.
To establish an A. cunicularia hypugaea target sequence, we amplified the target from 10 replicates of 10 ng burrowing owl blood DNA, sequenced each amplicon, and constructed a consensus sequence.
Testing Unfed Fleas Exposed to Animal Pelts
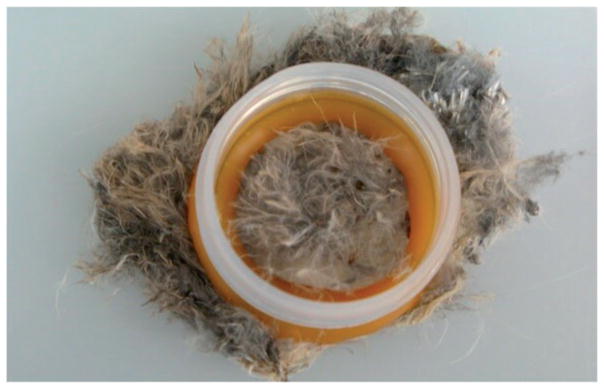
To simulate a scenario in which fleas infest a vertebrate host but do not feed on it, we exposed 27 newly emerged (unfed), colony-reared X. cheopis to a colony-reared mouse (AG129) pelt and 27 to a piece of burrowing owl pelt. Both pelts had been removed from frozen carcasses and stored at −20°C. They were allowed to thaw at ambient temperature immediately before the experiment. We attached a capsule to each pelt by cutting off the top portion of a 50-ml conical tube and securing it with a mixture of hot wax and resin. The area inside the capsule remained covered with fur or feathers. We then placed the fleas in the capsules and secured the conical tube lids so that the fleas were contained on the pelts (Fig. 2). After a 1-h exposure, we collected all the fleas and stored them at −20°C. We also collected pieces of skin from inside each capsule and stored these samples at −20°C. The fleas were later rinsed with DPBS and examined via light microscopy to verify that the exoskeleton was intact. They were then preserved at −20°C pending DNA isolation.
Fig. 2.
Fleas inside a capsule on a pelt from a burrowing owl nestling. To simulate a scenario in which fleas infest a vertebrate host but do not feed on it, unfed Xenopsylla cheopis were secured on owl or mouse pelts in capsules with screw-top lids (not pictured) for a 1-h exposure period. Fleas were subsequently tested using our bloodmeal assay to determine if the assay would detect vertebrate DNA.
We isolated and amplified DNA from each set of fleas, sequenced the amplicons, and analyzed the sequences using the same protocol we used for the field-collected fleas. As a positive control, each batch of extractions included a colony-reared X. cheopis that had been frozen immediately after feeding on a colony-reared SKH1 mouse. We also amplified and sequenced DNA extracted from the mouse and owl skin tissue using the DNeasy Blood and Tissue Kit (Qiagen, Valencia, CA). Animal procedures were approved by the Centers for Disease Control and Prevention Division of Vector-Borne Infectious Diseases Institutional Animal Care and Use Committee.
Results
Flea Identification
Of 81 fleas collected, 6 were too damaged to identify to genus. All of the remaining 75 fleas were identified as Pulex: 36 males and 39 females. Four of the males were too damaged to identify to species. We identified the remaining males (n =32) as P. irritans. We could not distinguish between P. irritans and Pulex simulans Baker females based on morphology, but because all identified males were P. irritans, all females were also presumed to be P. irritans.
Burrowing Owl Target Sequence
Using DNA extracted from burrowing owl whole blood, we established a 102-nt target sequence for this species. This consensus sequence matched the only A. cunicularia 12S sequence in the NCBI database (AF231330.1:27–119), except that our sequence included a 9-nucleotide insertion (CGAGCACTA) between nucleotides 69 and 70. Basic Local Alignment Search Tool (BLAST) analysis indicated that our 102-nt A. cunicularia sequence did not match any other sequence in the NCBI nucleotide collection; homologous sequences from other species differed by at least 9 nucleotides. We therefore considered samples yielding amplicons with this sequence positive for burrowing owl DNA.
Bloodmeal Identification
Of the 75 fleas identified as Pulex irritans or Pulex sp., 25 were too damaged to test using our bloodmeal assay. Of the 50 fleas we tested, 12 had no detectable vertebrate bloodmeal. The remaining 38 (76%) contained burrowing owl DNA.
Testing Unfed Fleas Exposed to Animal Pelts
All of the mouse-fed control flea samples and both the mouse and owl pelt samples to which the fleas had been exposed tested positive for vertebrate DNA. BLAST analysis indicated that the mouse skin amplicon sequence was consistent with the corresponding sequence for Mus musculus (KC663621.1:516-616). The owl skin amplicon sequence was identical to our consensus sequence from burrowing owl whole blood DNA. We did not detect vertebrate DNA in any of the unfed, pelt-exposed fleas (n =54).
Discussion
We found that a molecular assay developed to identify vertebrate bloodmeals in Ugandan cat fleas was flexible enough to detect burrowing owl DNA in North American P. irritans. We further demonstrated that the assay did not detect vertebrate DNA in fleas that had not taken a vertebrate bloodmeal but had been exposed to mouse or owl pelts. The assay is therefore unlikely to detect DNA in fleas from vertebrates that have served exclusively as phoretic hosts. Using this assay, we detected burrowing owl DNA in 38 P. irritans collected from burrowing owls in southern Idaho. We conclude that P. irritans feeds on burrowing owls.
Our finding raises questions about the potential effects of ecto-parasitism on owls. The few available studies of fleas on burrowing owls indicate that 1) Juveniles infested with fleas disperse from natal areas significantly later than those treated with insecticide, but only in some years (V. Garcia and C. Conway, unpublished data), 2) Nests with higher flea loads have lower productivity (J. Welty and J.R.B., unpublished data), and 3) Nest-site reuse does not appear to be affected by the presence or absence of fleas (Riding and Belthoff 2015). Other important aspects of the host–parasite relationship between burrowing owls and P. irritans are poorly understood, e.g., if and how this relationship alters owl physiology, immunology, parental care, or juvenile survival after leaving nests, and if it has any long-term reproductive consequences for the owls (e.g., Richner and Tripet 1999). Given that blood feeding depletes nutrients and can induce energy-demanding immune responses in the host (Møller et al. 2005, Krasnov 2008), our finding suggests that additional studies are merited to determine the extent to which P. irritans impact declining burrowing owl populations.
The finding that burrowing owls serve as a bloodmeal source for P. irritans also has potential implications for enzootic plague dynamics. Avian hosts are generally considered resistant to Y. pestis infection (Meyer 1950), although the wheatear (Oenanthe spp.), has been found to be infected with Y. pestis in the former Soviet Union and Mongolia (Shevchenko et al. 1980, Galdan et al. 2010). A vertebrate host must develop a high bacteremia (≥106 colony forming units/ml) to reliably infect feeding fleas (Engelthaler et al. 2000, Lorange et al. 2005). If an infected P. irritans feeds on a burrowing owl, it is unlikely that the burrowing owl will develop a sufficient bacteremia to subsequently infect other fleas. Also, taking an uninfected bloodmeal can reduce the probability that an infected flea transmits Y. pestis the next time it feeds (Eisen et al. 2006, 2007; Wilder et al. 2008a, b). Thus, if P. irritans serves as a Y. pestis vector within a landscape shared by burrowing owls and Y. pestis-susceptible mammals, P. irritans feeding on burrowing owls may actually reduce the force of infection within that ecosystem.
It is important to note, however, that in the Pacific Northwest, the geographic region where P. irritans most commonly parasitize burrowing owls, the owls are unlikely to have contact with mammals carrying infected P. irritans because the other primary P. irritans hosts are various Canidae (Hopla 1980). Canids, while susceptible to plague, rarely succumb to infection and are considered unlikely to provide a source of infection for feeding fleas (Gage et al. 1994). Burrowing owls in Idaho, Oregon, and Washington often nest in burrows dug by American badgers (Taxidea taxus) (Belthoff et al. 2015). Serologic surveys in this region indicate that the American badger has exposure to Y. pestis, but it develops only a transient infection (Messick et al. 1983). Thus, though there may be interaction between owls and some carnivores that are carrying fleas, those carnivores are unlikely to infect fleas and thus are unlikely to serve as a source of infected P. irritans. In this region, ground squirrels are the mammals most likely to develop a transmissible bacteremia. These species almost never host P. irritans (Hopla 1980, Barnes 1982).
Acknowledgments
We thank J.A. Montenieri and K. MacMillan for assistance with flea identification, J. Wade and G. Frye for assistance with field work, and the Idaho Bureau of Laboratories for assistance with shipping fleas to the CDC. We thank the U.S. Fish and Wildlife Service, Region 1 Avian Health and Disease Surveillance Monitoring Program, and the U.S. Bureau of Land Management, Morley Nelson Snake River Birds of Prey National Conservation Area, for logistical support.
References Cited
- Barnes AM. Surveillance and control of bubonic plague in the United States. Symp Zool Soc London. 1982;50:237–270. [Google Scholar]
- Belthoff JR, King RA. Nest-site characteristics of burrowing owls in the Snake River Birds of Prey National Conservation Area, Idaho. West North Am Nat. 2002;62:112–119. [Google Scholar]
- Belthoff JR, Bernhardt SA, Ball CL, Gregg M, Johnson DH, Ketterling R, Price E, Tinker JK. Burrowing owls, Pulex irritans, and plague. Vector Borne Zoonotic Dis. 2015;15:556–564. doi: 10.1089/vbz.2015.1772. [DOI] [PubMed] [Google Scholar]
- Blanc G, Baltazard M. Recherches expérimentales sur la peste; l’infection le la puce de l’homme: Pulex irritans L. Maroc Med. 1941;217:81–82. [Google Scholar]
- Boughton RK, Atwell JW, Schoech SJ. An introduced generalist parasite, the sticktight flea (Echidnophaga gallinacea), and its pathology in the threatened Florida scrub-jay (Aphelocoma coerulescens) J Parasitol. 2006;92:941–948. doi: 10.1645/GE-769R.1. [DOI] [PubMed] [Google Scholar]
- Brown JH. Sylvatic plague: The recovery of fleas from the burrowing owl and its burrow in a plage area in Alberta. Entomol News. 1944;55:15–18. [Google Scholar]
- Cantarero A, López-Arrabé J, Redondo AJ, Moreno J. Behavioural responses to ectoparasites in pied flycatchers Ficedula hypoleuca: An experimental study. J Avian Biol. 2013;44:591–599. [Google Scholar]
- Christe P, Richner H, Oppliger A. Of great tits and fleas: Sleep baby sleep. Anim Behav. 1996;52:1087–1092. [Google Scholar]
- Drancourt M, Houhamdi L, Raoult D. Yersinia pestis as a telluric, human ectoparasite-borne organism. Lancet Infect Dis. 2006;6:234–241. doi: 10.1016/S1473-3099(06)70438-8. [DOI] [PubMed] [Google Scholar]
- Eisen RJ, Lowell JL, Montenieri JA, Bearden SW, Gage KL. Temporal dynamics of early-phase transmission of Yersinia pestis by unblocked fleas: Secondary infectious feeds prolong efficient transmission by Oropsylla montana (Siphonaptera: Ceratophyllidae) J Med Entomol. 2007;44:672–677. doi: 10.1603/0022-2585(2007)44[672:tdoeto]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Eisen RJ, Bearden SW, Wilder AP, Montenieri JA, Antolin MF, Gage KL. Early-phase transmission of Yersinia pestis by unblocked fleas as a mechanism explaining rapidly spreading plague epizootics. Proc Natl Acad Sci USA. 2006;103:15380–15385. doi: 10.1073/pnas.0606831103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelthaler DM, Hinnebusch BJ, Rittner CM, Gage KL. Quantitative competitive PCR as a technique for exploring flea-Yersina pestis dynamics. Am J Trop Med Hyg. 2000;62:552–560. doi: 10.4269/ajtmh.2000.62.552. [DOI] [PubMed] [Google Scholar]
- Furman DP, Catts EP. Manual of medical entomology. 4. Cambridge University Press; Cambridge: 1982. Order siphonaptera; pp. 138–157. [Google Scholar]
- Gage KL, Montenieri JA, Thomas RE. The role of predators in the ecology, epidemiology, and surveillance of plague in the United States. Proceedings, Sixteenth Vertebrate Pest Conference, University of Nebraska; Lincoln. Davis: University of California; 1994. [Google Scholar]
- Galdan B, Baatar U, Molotov B, Dashdavaa O. Plague in mongolia. Vector Borne Zoonotic Dis. 2010;10:69–75. doi: 10.1089/vbz.2009.0047. [DOI] [PubMed] [Google Scholar]
- Graham CB, Black WCI, Boegler KA, Montenieri JA, Holmes JL, Gage KL, Eisen RJ. Combining real-time polymerase chain reaction using SYBR Green I detection and sequencing to identify vertebrate blood meals in fleas. J Med Entomol. 2012;49:1442–1452. doi: 10.1603/me12071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham CB, Borchert JN, Black WCI, Atiku LA, Mpanga JT, Boegler KA, Moore SM, Gage KL, Eisen RJ. Blood meal identification in off-host cat fleas (Ctenocephalides felis) from a plague-endemic region of Uganda. Am J Trop Med Hyg. 2013;88:381–389. doi: 10.4269/ajtmh.2012.12-0532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz NG. Rodent reservoirs and flea vectors of natural foci of plague. In: Dennis DT, Gage KL, Gratz N, Poland JD, Tikhomirov E, editors. Plague manual: Epidemiology, distribution, surveillance and control. World Health Organization; Geneva, Switzerland: 1999. pp. 63–96. [Google Scholar]
- Hopla CE. A study of the host associations and zoogeography of Pulex. In: Traub R, Starcke H, editors. Proceedings, International Conference on Fleas; 1977; Peterborough, UK. Rotterdam: A.A. Balkema; 1980. pp. 185–207. [Google Scholar]
- Jellison WL. Sylvatic plague: Studies of predatory and scavenger birds in relation to its epidemiology. Public Health Rep. 1939;54:792–798. [Google Scholar]
- Klute DS, Ayers LW, Green MT, Howe WH, Jones SL, Shaffer JA, Sheffield SR, Zimmerman TS. Status assessment and conservation plan for the western burrowing owl in the United States. U.S. Department of Interior; Fish and Wildlife Service, Biological Technical Publication; Washington, DC: 2003. FWS/BTP-R6001-2003. [Google Scholar]
- Krasnov BR. Functional and evolutionary ecology of fleas: A model for ecological parasitology. Cambridge University Press; New York, NY: 2008. [Google Scholar]
- Laudisoit A, Leirs H, Makundi RH, Van Dongen S, Davis S, Neerinckx S, Deckers J, Libois R. Plague and the human flea, Tanzania. Emerg Infect Dis. 2007;13:687–693. doi: 10.3201/eid1305.061084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis RE, Lewis JH, Maser C. The fleas of the Pacific Northwest. Oregon State University Press; Corvallis, OR: 1988. [Google Scholar]
- Lorange EA, Race BL, Sebbane F, Hinnebusch BJ. Poor vector competence of fleas and the evolution of hypervirulence in Yersinia pestis. J Infect Dis. 2005;191:1907–1912. doi: 10.1086/429931. [DOI] [PubMed] [Google Scholar]
- Messick JP, Smith GW, Barnes AM. Serologic testing of badgers to monitor plague in southwestern Idaho. J Wildl Dis. 1983;19:1–6. doi: 10.7589/0090-3558-19.1.1. [DOI] [PubMed] [Google Scholar]
- Meyer KF. Immunity in plague; a critical consideration of some recent studies. J Immunol. 1950;64:139–163. [PubMed] [Google Scholar]
- Møller AP, Christe P, Garamszegi LZ. Coevolutionary arms races: Increased host immune defense promotes specialization by avian fleas. J Evol Biol. 2005;18:46–59. doi: 10.1111/j.1420-9101.2004.00774.x. [DOI] [PubMed] [Google Scholar]
- Ratovonjato J, Rajerison M, Rahelinirina S, Boyer S. Yersinia pestis in Pulex irritans fleas during plague outbreak, Madagascar. Emerg Infect Dis. 2014;20:1414–1415. doi: 10.3201/eid2008.130629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richner H, Tripet F. Ectoparasitism and the trade-off between current and future reproduction. Oikos. 1999;86:535–538. [Google Scholar]
- Richner H, Oppliger A, Christe P. Effect of an ectoparasite on reproduction in great tits. J Anim Ecol. 1993;62:703–710. [Google Scholar]
- Riding CS, Belthoff JR. Removal of old nest material decreases reuse of artificial burrows by burrowing owls. Wildl Soc Bull. 2015;39:521–528. [Google Scholar]
- Shevchenko VL, I, Grigor’ev S, Altukhov AA, Mashtakov VI. Case of the isolation of the causative agent of plague (Yersinia pestis) from the Isabelline wheatear (Oenanthe isabellina Temm.) Med Parazitol (Mosk) 1980;49:85–86. [PubMed] [Google Scholar]
- Skoruppa MK, Pearce B, Woodin MC, Hickman GC. Ectoparasites of burrowing owls (Athene cunicularia hypugaea) wintering in southern Texas. Tex J Sci. 2006;58:73–78. [Google Scholar]
- Smith BW, Belthoff JR. Identification of ectoparasites on burrowing owls in southwestern Idaho. J Raptor Res. 2001;35:159–161. [Google Scholar]
- Verjbitski DT. XXVI. The part played by insects in the epidemiology of plague. J Hyg. 1908;8:162–208. doi: 10.1017/s0022172400003260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler CM, Douglas JR, Evans FC. The role of the burrowing owl and the sticktight flea in the spread of plague. Science. 1941;94:560–561. doi: 10.1126/science.94.2450.560. [DOI] [PubMed] [Google Scholar]
- Wilder AP, Eisen RJ, Bearden SW, Montenieri JA, Gage KL, Antolin MF. Oropsylla hirsuta (Siphonaptera: Ceratophyllidae) can support plague epizootics in black-tailed prairie dogs (Cynomys ludovicianus) by early-phase transmission of Yersinia pestis. Vector Borne Zoonotic Dis. 2008a;8:359–367. doi: 10.1089/vbz.2007.0181. [DOI] [PubMed] [Google Scholar]
- Wilder AP, Eisen RJ, Bearden SW, Montenieri JA, Tripp DW, Brinkerhoff RJ, Gage KL, Antolin MF. Transmission efficiency of two flea species (Oropsylla tuberculata cynomuris and Oropsylla hirsuta) involved in plague epizootics among prairie dogs. Ecohealth. 2008b;5:205–212. doi: 10.1007/s10393-008-0165-1. [DOI] [PubMed] [Google Scholar]