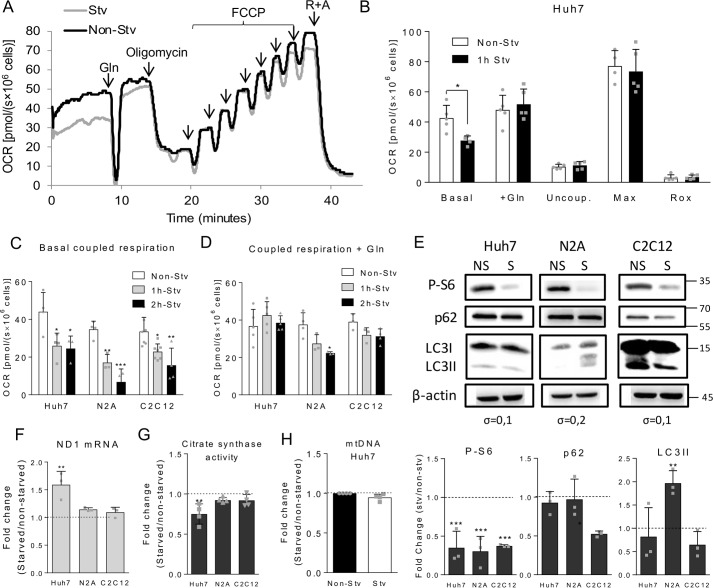
Figure 1.
Effects of short-term nutrient deprivation on Huh7, N2A, and C2C12 cells. A, representative respirometry experiment using Huh7 cells subjected or not to 1-h starvation (Stv or Non-Stv, respectively) followed by reintroduction of 2.5 mm glutamine in the culture medium during OCR recording. After OCR stabilization, the following ETS modulators were added: 250 nm oligomycin to measure uncoupled respiration, sequential additions of FCCP to achieve maximum respiration (at 1 mm), and 0.5 mm rotenone and 3.65 μm antimycin (R+A) to determine Rox rates. B, OCR recordings of non-starved or starved Huh7 cells in DMEM-A in the following conditions: basal respiration, after addition of 2.5 mm glutamine (+Gln), after inhibition of ATP synthase with oligomycin (uncoupled respiration (Uncoup.), upon titration with FCCP (maximum respiration (Max)), and after inhibition of the respiratory complexes with antimycin and rotenone (Rox). C, coupled respiration (basal respiration minus uncoupled respiration values) of Huh7, C2C12, and N2A cells subjected to 1- and 2-h starvation. D, coupled respiration after glutamine reintroduction to non-starved, 1-, or 2-h-starved Huh7, N2A, or C2C12 cell suspensions. E, immunodetection by Western blotting of P-S6, p62, LC3I, LC3II (the cleaved form of LC3I), and β-actin in Huh7, N2A, and C2C12 cell lysates in non-starved (NS) and starved (S) conditions. The position of the molecular weight markers are indicated on the left, and the standard deviations of the differences between non-starved and starved conditions are shown below the β-actin blotting. The lower panels show the -fold change observed for the densitometric quantification of the bands between starved and non-starved conditions for p62, LC3II, and P-S6. F, relative mRNA expression of the mitochondrial gene ND1 between starved and non-starved Huh7, N2A, and C2C12 cells. G, relative activity of citrate synthase between starved and non-starved Huh7, N2A, and C2C12 cells. H, relative quantification of mitochondrial DNA between starved and non-starved Huh7 cells. For each graph, individual data points are represented with symbols. Error bars represent mean ± S.D., and asterisks indicate significant differences between non-starved and starved conditions for each cell line: *, p < 0.05; **, p < 0.01; ***, p < 0.001.