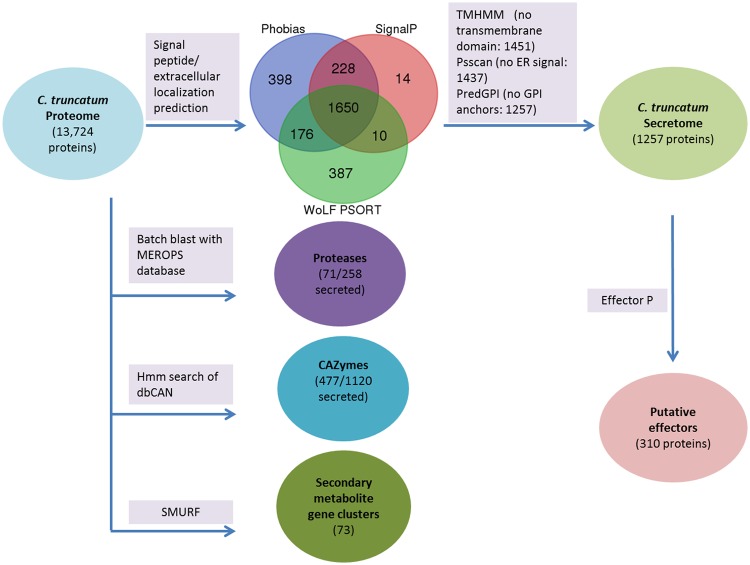
Fig 3. Work flow for functional annotation of some of the gene categories relevant to pathogenicity.
A stringent pipeline of tools was used to predict 1257 proteins that are highly likely to be secreted and other important categories like carbohydrate active enzymes (CAZymes), proteases and secondary metabolism (SM) gene clusters. 310 of the secreted proteins were predicted to be putative effectors, 477 were secreted CAZymes and 71 were secreted proteases. Secondary metabolite backbone genes were not detected in the secretome.