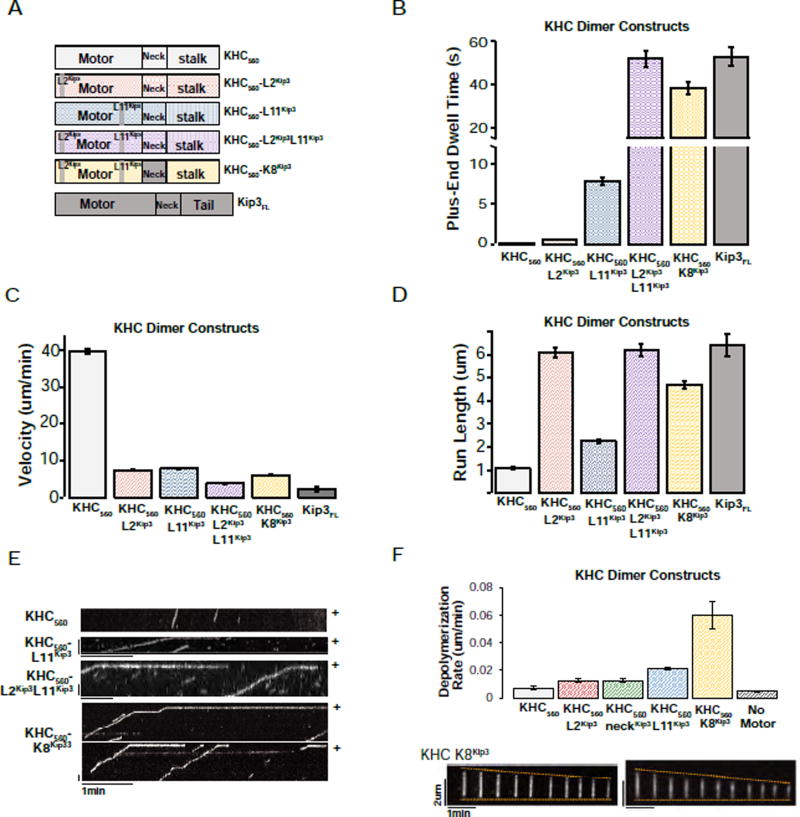
Figure 3. Kip3’s L11 is sufficient to convey plus-end binding to KHC and Kip3’s L2 is sufficient to increase the processivity of KHC.
A. Domain swaps of Kip3 segments into dimeric KHC. Diagrams of the chimeric constructs used. Superscripts denote the origin of the inserted segment.
B. Domain swap showing that Kip3 Loop11 is a transportable segment sufficient for prolonged plus-end binding. Kip3 and a KHC chimera containing Kip3’s L2, L11 and the family-specific neck segment (KHC560-K8Kip3) have comparable plus-end residence times. Shown is the end-residence time (mean +/− SEM; n=149–394 events) of the indicated KHC variant constructs compared to unmodified KHC560 and Kip3FL. The superscript denotes the origin of the indicated loop that was grafted into KHC560.
C. Either Kip3 L2 or L11 reduce the velocity of KHC560. The velocity (mean +/− SEM; n=155–795 events) of KHC560, KHC chimeras and Kip3 on taxol-stabilized microtubules.
D. Kip3 and a KHC chimera containing Kip3’s L2, L11 and family-specific neck segment (KHC560-K8Kip3) have comparable run lengths (mean +/− SEM; n=155–795 events.)
E. Representative kymographs from single molecule assays illustrating the motility and plus-end residence of KHC560 and KHC560 variant motor constructs. Scale bar: 1 min (horizontal), 1 um (vertical).
F. KHC-K8Kip3 is a synthetic microtubule depolymerase. Depolymerization rate of GMPCPP-stabilized microtubules by the indicated kinesin-1 chimeras (120nM, mean +/− SEM; nMT=72–278). The KHC560-NeckKip3 construct has a velocity of 7.4 +/− 0.1 um/min and a run length of 1.2 +/− 0.1 (n=547), similar to KHC. Bottom: GMPCPP-stabilized microtubules shown at 29s intervals after the addition of 120 nM KHC560-K8Kip3 (nMT=84). Scale bars: 2 um (vertical), 1 min (horizontal).