Abstract
Cystic Fibrosis (CF) is the most common monogenic disease among people of Western European descent and caused by mutations in the CFTR gene. However, the disease severity is immensely variable even among patients with similar CFTR mutations due to the possible effect of ‘modifier genes’. To identify genetic modifiers, we applied RNA-seq based transcriptomic analyses in CF patients with a mild and severe lung phenotype. Global gene expression and enrichment analyses revealed that genes of the type I interferon response and ribosomal stalk proteins are potential modifiers of CF related lung dysfunction. The results provide a new set of CF modifier genes with possible implications as new therapeutic targets for the treatment of CF.
Introduction
Cystic fibrosis (CF) is the most common life-threatening genetic disease among people of Western European descent. The disease is caused by a mutation in the underlying disease-conferring gene, Cystic Fibrosis Transmembrane and Conductance Regulator (CFTR) that encodes a chloride channel. CFTR mutations result in disrupted chloride transport in the epithelial cells of various organs including lung, intestine, pancreas, and testes [1]. More than 2000 CFTR genetic variants have been reported and categorized into six classes, as these mutations were observed to exhibit diverse effects on the production of CFTR protein, its trafficking, its function, and its stability at the epithelial cell membrane [1, 2]. The most prevalent mutation in CF is the lack of a phenylalanine residue at position 508 (also known as F508del), which results in a misfolded CFTR protein with reduced transmembrane conductance of chloride ions [3]. Due to the trafficking defect, the F508del mutation prevents the CFTR protein reaching the apical membrane of epithelial cells and also interrupts the normal movement of chloride ions by disrupting CFTR channel gating [4].
Chronic progressive lung disease is the primary reason for morbidity and mortality in CF. However, the disease severity is immensely variable even among patients with similar CFTR mutations. For instance, CF patients with a homozygous F508del mutation showed varying phenotypic heterogeneity in terms of lung function that is measured by forced expiratory volume in 1 second (FEV1) [5]. It has been observed that CFTR genotypes per se do not corroborate with the pulmonary phenotype in unrelated CF patients [6]. However, twin and sibling studies clarified that the modifier genes and their genetics contributed substantially to divergent outcomes observed in CF lung disease [7, 8]. As CF is a multi-organ disease, the contribution of modifier genes towards disease severity was relatively higher when compared to non-genetic modifiers in specific organs [9].
Modifier genes are a vital part of human genetics and it is argued that they are responsible for the diverse phenotype observed with various diseases, more precisely in CF [10, 11]. Several CF related modifier genes have been reported through genome-wide association studies (GWAS), hypothesis-driven candidate gene studies, and microarray based transcriptomic analyses [5, 9, 12–15]. Unlike GWAS and candidate gene studies, transcriptomic analyses provide information on both genetic modifiers and non-genetic modifiers such as infections from the environment. The host immune system responds to these infections by activating immune genes, and these gene transcripts can be measured by transcriptomic approaches. Though microarray-based transcriptomic analyses could identify some modifier genes, the technology is limited to known transcripts [12, 16, 17]. However, global gene expression analysis using RNA-seq provides several advantages over microarray including the detection of a higher number of differentially expressed genes and the detection of novel transcripts [18]. Therefore, in the present study we performed a transcriptomic analysis using RNA-seq on CF patients with an identical genotype (homozygous F508del), but with mild and severe lung dysfunction determined by FEV1 measurements.
Materials and methods
Patient samples
A total of 32 F508del homozygous CF patients were recruited for the study from the University Childrens’ Hospital, Tuebingen, Germany and the Division of Paediatric Respiratory Medicine, University of Bern, Switzerland. All blood samples were collected in PaxGene blood vacutainer tubes to ensure RNA quality and significantly reduce RNA degradation. Participants were classified into severe CF patients (n = 16) and mild CF patients (n = 16) based on their FEV1 values, which is a standard to quantify the clinical phenotype of CF. The detailed clinical characteristics of the enrolled patients are described in Table 1. For statistical analyses of clinical parameters, data were analyzed using the Mann-Whitney U test and are presented as means and SDs, unless stated otherwise. Ethical approval was obtained from the Institutional review board, Children’s University Hospital Tuebingen, Germany. Informed written consent was obtained from all participants; for those who were children, consent was obtained from respective parents or guardians.
Table 1. Clinical characteristics of the mild CF and severe CF samples.
| Clinical Parameters | Mild CF patients | Severe CF patients | P value |
|---|---|---|---|
| Number (n) | 16 | 16 | Not Significant |
| Age, mean (SD) | 25 (12) | 21 (9) | Not Significant |
| Male:Female | 10:6 | 8:8 | Not Significant |
| CFTR Mutation | F508del/ F508del | F508del/F508del | Not Significant |
| FEV1 (% predicted), mean (SD) | 92 (16) | 36 (10) | < 0.0001*** |
| Infections | PSA, SA, CA, SM | PSA, SA, CA, SM, AF | Not Significant |
*** Mann-Whitney U test.
PSA, Pseudomonas aeruginosa; SA, Staphylococcus aureus; CA, Candida albicans; SM, Stenotrophomonas maltophilia; AF, Aspergillus fumigatus.
RNA library preparation
Total RNA was isolated with PAXgene tubes using a Qiacube roboter with standard protocols (www.qiagen.com). Excess globin mRNA was removed with GLOBINclearTM kit and sequencing libraries were prepared with the TruSeqTM Total RNA Sample Prep Kit following the manufacturer’s instructions (www.illumina.com). Each sequencing library was tagged with a 6 nucleotide long barcode that identifies from which sample a sequence was derived. After size selection, all sequencing libraries were quantified on a Qubit® fluorometer and equimolar amounts of 16 libraries were combined to generate two sequencing pools. These were loaded onto 8 lanes of an Illumina GAIIx single-read flow cell and two MiSeq flow cells. Bound molecules were clonally amplified on a cBot instrument. Subsequently, the first 50 nucleotides from each fragment were sequenced followed by a seven nucleotide sequencing run to decipher the barcode sequence in the adapter (www.illumina.com).
RNA-seq data analysis
The quality of raw sequenced reads was assessed using the fastqc quality control tool and high-quality reads were aligned against the Homo sapiens reference genome (hg19) using the STAR RNA-seq alignment tool [19]. The STAR aligners automatically detect and remove adaptor sequences from the sequence reads before alignment. Next, we calculated the total number of mapped reads and the number of uniquely mapped reads for each sample. As an additional layer of quality control, we discarded samples with low mapped read count and samples with a small percentage of uniquely mapped reads. For samples that pass the additional quality check, we estimated the gene expression level as CPM (count per million mapped reads) using HTSeq [20]. We used EdgeR R package to identify differentially expressed genes (DEGs) between mild CF and severe CF [21]. RNA-Seq reads are available in NCBI-Sequence Read Archive (SRA) database, under accession number SRP111640. Samples in each CF group were treated as biological replicates and only genes that pass the significance cut-off were treated as DEGs. In this study, we used log fold-change in expression less than -0.5 or greater than 0.5 and P-value (FDR corrected P-value) less than 0.05 as the significance cut-off to identify DEGs. In order to functionally categorize DEGs, we tested enrichment of Gene Ontology (GO) terms among DEGs using Amino.2 (www.amigo.geneontology.org). Additionally, we also analyzed DEGs for the over-representation of KEGG pathways (http://www.genome.jp/kegg/pathway.html). Motif activity response analysis (MARA) was performed to predict the global regulatory interaction of RNA-seq data using ISMARA online tool (www.ismara.unibas.ch). MARA predicts transcription factor (TF) binding sites with a specific algorithm and calculates the influence of specific TF in terms of gene expression in a given sample. Mann-Whitney U rank sum tests were applied to analyze differences in motif activity between the study groups. All the major DEGs were checked for their association with CF using Open target Platform (www.targetvalidation.org).
qPCR validation
qRT-PCR validation was performed for 16 of the DEGs utilizing the same samples utilized for RNA-seq as a technical reproducibility operating different platforms. Primers were designed with the Primer3 online tool (www.primer3.ut.ee) Primers, amplicon details, and cycling conditions are listed in Table 2. cDNA was synthesized using iScript (www.biorad.com), followed by qRT-PCR using Power SYBR green, and samples were run in triplicate on a ViiA7 Real-Time PCR System (www.lifetechnologies.com). The relative expression levels of selected DEGs were normalized to the 18S rRNA. Differences in mRNA expression between mild CF and severe CF were analyzed by pair-wise fixed reallocation randomization tests with REST 2009 software [22]. The severe CF patients group was used as a control group and statistical significance was determined using randomization tests. In addition, the expression of each gene was determined for every single patient using the 2-ΔΔCt method and Mann-Whitney U tests were applied for separate gene analyses between study groups. The level of significance was set to a P-value of <0.05.
Table 2. Primer details and PCR cycling conditions for the qPCR validation.
| Gene | Primer sequences (5'-3') | Amplicon Size (bp) | Annealing Temperature (°C) |
|---|---|---|---|
| HERC5 | F: CCAGCTTGCTTGTCCAACAG | 157 | 58 |
| R: CGGCCAGTAAACCCTCTTCT | |||
| IFIT1 | F: TGGACCCTGAAAACCCTGAA | 243 | 54 |
| R: TCTGTGAGGACATGTTGGCT | |||
| IFIT2 | F: GCGAAACAACTGCTCCATCT | 205 | 55 |
| F: CCAAGACATGCAAAGCCTCA | |||
| RSAD2 | F: AAGAGGAGGAAGAGGACCCT | 250 | 55 |
| R: CAGAACCTCACCAACTTGCC | |||
| IFI44L | F: GAGCAACTGGTGTGTCGTTT | 213 | 56 |
| F: CCTATTTCTGTGCTCTCTGGC | |||
| GOS2 | F: GGAATGGAGAGACAGAGGGG | 239 | 59 |
| R: AGTGCAAAATGGTAGACGCA | |||
| CXCL10 | F: TGGATGTTCTGACCCTGCTT | 201 | 56 |
| R: AAAGAATTTGGGCCCCTTGG | |||
| FOSB | F: TCTGTCTTCGGTGGACTCCTTC | 209 | 57 |
| R: GCAAACCGTAGATGCTCAGGG | |||
| OAS3 | F: GCTTCACAGAGCTACAACGG | 167 | 58 |
| R: CTCCCAGGCATACACAGTCA | |||
| IFI6 | F: AGCAGCGTCGTCATAGGTAA | 213 | 56 |
| R: TGCACTCTAGCCTGGACAAT | |||
| MX1 | F: CATCCAGCCACCATTCCAAG | 168 | 57 |
| R: AGAATCGCTTGAACCTGGGA | |||
| EGR1 | F: AGCTGGAGGAGATGATGCTG | 257 | 54 |
| R: CCAGCACCTTCTCGTTGTTC | |||
| IL8 | F: TCTTGGCAGCCTTCCTGATT | 211 | 56 |
| R: TCCAGACAGAGCTCTCTTCCATC | |||
| LOC644172 | F: CCGACGTCCATTTCTCCAAG | 184 | 58 |
| R: TCATCCACTTCCAGCTCAGG | |||
| ZFN683 | F: AGCCTTGCCTTACCCGCTGAAA | 129 | 60 |
| R: AATGGACGCTCTCCACTGTGCA | |||
| EPB41L4B | F: ACCCACTTCCTTGACAGAGT | 233 | 55 |
| R: CGCAAGTTAGCAGCACCAAT | |||
| 18s rRNA | F: GTAACCCGTTGAACCCCATT | 151 | 54 |
| R: CCATCCAATCGGTAGTAGCG | |||
Results
We investigated the transcriptomic profile of peripheral blood leukocytes from mild and severe CF patients using RNA-sequencing. No differences were evident for important clinical parameters between the investigated groups, except for the FEV1 status (P<0.0001; Table 1). After the quality check for RNA-seq data, three samples were removed from downstream analyses due to the low number of total reads and lower mapping rate (S1 Fig). In total, 12,778 genes with at least ten reads per sample were included in the final analysis stage. The principal component analysis (PCA) of the final data revealed that the differences in the expression profiles of the two investigated groups were relatively small (S2 Fig).
Differentially expressed genes (DEGs) between mild CF and severe CF
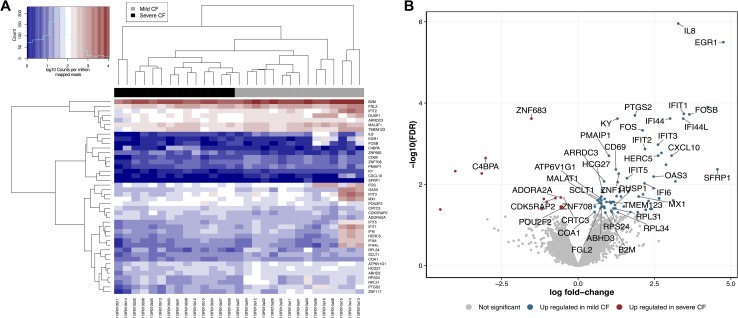
Differential expression analysis showed that 88 genes were differentially expressed between mild and severe CF group of patients, among which 74 genes (84%) had higher expression levels in mild CF patients and 14 (16%) genes had higher expression levels in individuals with severe CF. The results from this analysis with respective expression levels and adjusted P-values can be found in S1 Table. We compared DEGs between mild and severe CF patients and the heat-map of the selected DEGs revealed two distinct clusters for the investigated groups (Fig 1A). Global gene level expression analysis between the groups (mild CF vs. severe CF) is shown in volcano plots (Fig 1B) with respective gene name, expression level and accuracy of the detection using RNA-seq. EGR1, SFRP1, RSAD2, and FOSB are the major DEGs that were significantly overexpressed (>3.5) in the mild CF group in comparison with the severe CF group. Increased expression of EGR1 was found to be significantly associated with mild lung disease (fold change = 4.5; FDR P = 3.1x10-6). Concordantly, other EGR family genes, EGR2 (fold change = 2.4; FDR P = 0.04) and EGR3 (fold change = 2.6; FDR P = 0.02) showed higher level of expression in the mild CF group. Similarly, EPB41L4B, LOC644172, C4BPA, and ZNF683 are the major DEGs that were significantly overexpressed (>1.5) in the severe CF group (S1 Table). Among these genes, ZNF683 exhibited better association with the severe form of CF lung disease (fold change = 1.5; FDR P = 0.0002).
Fig 1. RNA-seq analyses of Mild CF and severe CF patients.
A) Heat-map of differentially expressed genes. Each column represents a separate patient, and each horizontal line represents a separate gene. Dendrogram of clustered samples and genes, in which mild CF and severe CF samples cluster with respect to their expression similarity. Expression profiles are measured by counts per million reads (CPM- model). B) Volcano plot of RNA-seq data, in which the -Log10 of the false discovery rate is plotted against Log2 fold change.
qPCR validation of RNA-seq data
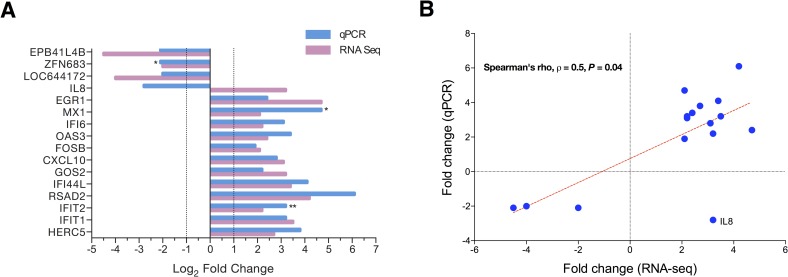
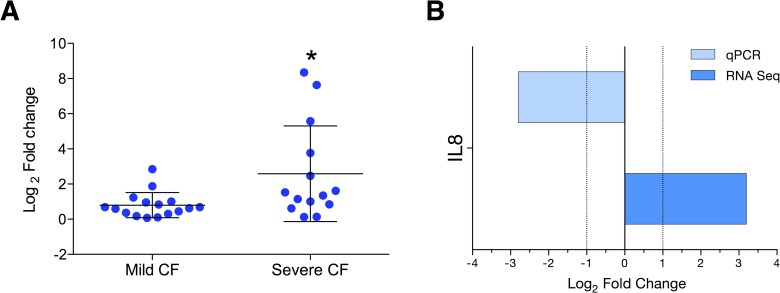
We validated RNA-seq findings for 16 DEGs by qPCR using the same samples and observed high concordant results between RNA-seq and qPCR (94%; 15 out of 16 DEGs were verified; Fig 2A). The comparison of DEG expression levels between RNA seq versus qPCR revealed better correlation (Spearman’s rho, ρ = 0.53, P = 0.04; Fig 2B). Thus, these 15 genes had similar mRNA levels, as RNA-Seq and qRT-PCR data were comparable. Furthermore, significant difference in expression levels was observed in qPCR results for the mild CF group with following genes, IFIT2 (fold change = 3.2, P<0.001), MX1 (fold change = 3.2, P<0.05) and ZFN683 (fold change = -2.1, P<0.05). No significant difference was achieved for EGR1 as many samples of the severe CF group failed in amplification (data not shown). IL-8 was the only DEG that could not be verified by qPCR. Since IL-8 is a very important proinflammatory cytokine in CF settings, we further dissected the IL-8 data for both platforms. RNA-seq data showed that IL-8 was overexpressed in the mild CF group (Figs 2A and 3B). Conversely IL-8 was significantly overexpressed in the severe CF group according to qPCR analysis (P = 0.01; Fig 3A).
Fig 2. qPCR validation of RNA-seq findings.
A) Expression profile of RNA-seq data and qPCR data for selected genes were compared using the same samples. Severe CF samples were used as control group and the expression level set to 1 or -1. Expression was normalized using 18s as a reference gene. Results represent mean values and are expressed as Log2 values of the fold change. Significant difference observed in qPCR results between mild CF and severe CF patients (**P value < 0.05; ***P value < 0.001). The level of significance was set to a P-value of < 0.0001 for RNA-seq data. B) Correlation analysis of RNA-seq and qPCR. (Log2 values of the fold change; Spearman’s rho, ρ = 0.53, P value = 0.04).
Fig 3. IL-8 expression by qPCR and RNA seq.
A) Dot-plot shows the expression of IL-8 in severe and mild CF patients. Expression was normalized using 18s as a reference gene. Results represent mean values and are expressed as Log2 values of the fold change. B) Using the same data of Fig 2A for IL-8 to understand different results between platforms.
Enrichment and MARA analysis
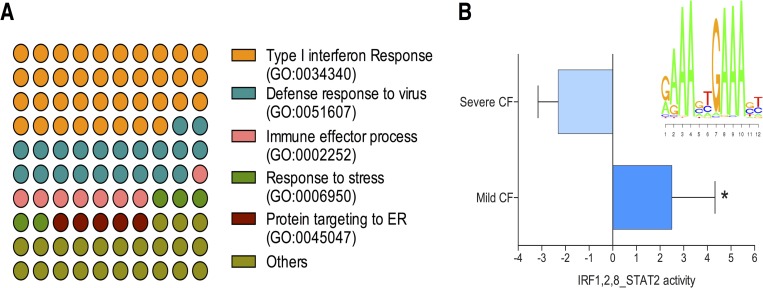
To understand the biological processes that could be modified differently between the mild CF and the severe CF group, we performed a GO enrichment analysis for DEGs that are upregulated in mild and severe CF. In case of the mild CF group, genes that belongs to type I interferon response were selectively enriched (38%, P = 1.45E-09; Fig 4A, S2 Table). A total of ten genes involved in this pathway showed elevated expression in the mild CF group (EGR1, MX1, IFIT1, IFIT2, IFIT3, OAS3, OASL, IFI6, ISG15, and RSAD2). In addition, the pathways of the anti-viral response and the immune effector process showed significant enrichment (S2 Table). Enrichment analysis identified three ribosomal stalk protein genes RPL31, RPL34, and RPS24 were enriched in the pathway of protein targeting to the endoplasmic reticulum (ER). No specific pathway was enriched for the fourteen-upregulated genes of the severe CF group. To explore the contribution of specific TFs and regulatory networks of DEGs we implemented MARA analysis to our RNA-seq data. Our data revealed that IRF1, IRF2, IRF8,and STAT2 transcription factor activity were significantly increased in the mild CF group (mean activation z-score for mild CF 2.5, mean activation z-score for severe CF -2.3; P < 0.05; Fig 4B) with elevated transcript levels of their regulated genes. Top target genes (Target score >10) for these TFs belong to the type I interferon response pathway. The highest score (Target score >25) was observed for the following genes: RSAD2, IFI44L, IFIT1, ISG15, IFIT3, OAS3, HERC5, and IFI44 (S1 File). Both our enrichment and pathway analysis were consistent in identifying the possible contribution of the type I interferon response in the mild CF phenotype. The network analysis of IRF1,2,8 and STAT2 transcription factors showed closer interactions of the above mentioned genes (S3 Fig) and other related genes in the type I interferon response.
Fig 4. Functional enrichment analysis of RNA-seq data.
A) Representative Gene Ontology (GO) terms enrichment among differentially expressed genes for biological processes. Genes involved in Type I interferon response, and ribosomal proteins responsible for endoplasmic reticulum transport and protein synthesis were over represented. B) ISMARA analysis predicts significant difference in IRF1,2,8 and STAT2 activity between mild CF and severe CF patients (*P value < 0.05).
Discussion
CF patients with identical inherited mutations in the CFTR locus exhibit substantial variation in disease severity and lung function. The phenotypic heterogeneity of CF is majorly affected by modifier genes [23]. Though several approaches are utilized to identify possible modifier genes of CF, here we used a transcriptomics approach for this purpose [11]. A previous study by Wright et al., attempted to identify modifier genes between mild and severe CF patients in their nasal respiratory epithelial cells by a transcriptomics approach using microarray technology. However global gene expression analysis using RNA-seq has many advantages over microarray [24]. Therefore, in this study we primarily used RNA-seq technology to identify modifier gene(s) of mild and severe lung phenotype in CF patients homozygous for the F508del mutation. Primarily, we assessed whether our transcriptomics results from peripheral blood leukocytes were comparable to native cells of the respiratory system (nasal, tracheal, and bronchial epithelial cells) to avoid possible difference in these cell types. We compared our data with six different studies and observed similar expression patterns for genes including CXCL10, GOS2, PTGS2, IFIT1, IFIT3, and ISG15 [12, 16, 17, 25–27]. Thus, upregulated gene profiles are relatively similar between blood leukocytes and native respiratory cells in CF. Likewise, both RNA-seq and microarray platforms resulted in common upregulated genes. Our data suggest that the blood cell transcriptome can be used as a surrogate for both upper and lower airway respiratory cells in CF. In addition, our RNA-seq data were validated using qPCR with the same samples and resulted in a high coherence score (94%).
The differences in the expression profiles of the two investigated groups are relatively small as they resulted in only 88 DEGs. However, we observed striking differences for these genes between the mild and severe pulmonary phenotype of CF. Seventy four genes exhibited higher expression in patients with mild CF lung disease compared with severe CF lung disease. A total of 14 genes showed a significant upregulation in individuals with severe CF compared with those with mild CF. Interestingly, enrichment analysis revealed that genes involved in the type I interferon response and the defense response to viral infections were highly enriched in the mild CF group. Though previous studies reported the association of type I interferon genes (IFIT1, IFIT3, and ISG15) with CF [12], the present study confirms the role of the type I IFN pathway in modifying CF. A very recent study using a systems biology approach identified a type I interferon gene IFI16, as a major CF modifier gene that alters lung function [28]. The type I IFNs are well-known for anti-viral response, and viral infections are potential contributors for a decline in lung function in CF patients [29]. Therefore, we carefully considered the influence of viral infections with the observed expression profile. However, all our study participants did not exhibit symptoms of cold or upper respiratory viral infections during blood sampling. Therefore, we deemed that the contribution of viral infections to the observed DEGs between mild and severe CF patients is less likely. However, increasing evidences suggest that Type I IFN signaling pathway is involved in host defense against other pathogens including bacteria, parasite and fungi [30]. For example, the type I interferon gene expression and their polymorphisms were prominently associated with susceptibility to systemic candidiasis [31]. Notably, earlier studies confirmed that the type I interferon signaling was activated by Pseudomonas aeruginosa in normal lung epithelial cells but was abrogated in CF epithelial cells [32]. P. aeruginosa is a well-known pathogen in CF patients that has been directly associated with decline in pulmonary function [33]. Of note, our study patients in both groups had fungal and bacterial infections without significant differences in their distribution between groups. Therefore, it is rational to hypothesize that due to the enhanced Type I IFN signaling, mild CF individuals kept their infections and lung function decline in control. This hypothesis can be further supported by the anti-bacterial effect of the type I IFN response in lung epithelium and the respiratory tract [34, 35]. The effect of another interferon related gene, IFRD1, in modifying the pathogenesis of CF is well studied by others and us [15, 36]. Together, our findings support the contribution of IFN, specifically, the type I IFN response in modifying lung function associated with CF.
Among the upregulated genes of the type I IFN response, EGR1 was a prominent gene that shows higher expression in mild CF patients. A previous transcriptomic study had identified that EGR1 was downregulated in P. aeruginosa infected CF bronchial epithelial cells [37]. Besides, EGR1 was observed to be upregulated in A. fumigatus and Toxoplasma gondii infected cells [38], which signifies its significance during infections. Interestingly, we found that two other EGR family members, EGR2 and EGR3, were also upregulated in the mild CF group. EGRs are a family of DNA-binding zinc-finger proteins and function as transcriptional regulators. In addition, EGR proteins were found to closely interact with other type I IFN genes including Mx1, HERC5, RSAD, and others. Thereby, the role of EGRs as potential modifier genes in CF lung disease warrants future research. Other potential modifier genes in type I IFN signaling are Mx1 and IFIT2, as they play a dominant role against P. aeruginosa and their expression also was verified by qPCR [35]. MARA analysis resulted in higher transcriptional activity of IRF1,2,8 and STAT2 TF in mild CF patients and they are responsible for the higher expression of Type I IFN genes. Enrichment analysis identified three ribosomal stalk proteins RPL31, RPL34 and RPS24, that were highly expressed in individuals with mild CF. These proteins were observed to be involved in protein trafficking to the ER and golgi transport. The relevance of ribosomal stalk proteins in CF is well documented. Earlier study in primary human bronchial epithelial cells with a homozygous F508del mutation showed that silencing of RPL12 was rescued by CFTR ion channel activity [39]. In addition, another ribosomal stalk protein RPL27 was reported as chloride-dependent gene, which expression is related to the function of the CFTR channel [40]. As the F508del mutation is related to protein trafficking and early degradation in ER, these ribosomal stalk proteins are attractive for further research.
The CF lung environment is dominated by neutrophils and elevated levels of the proinflammatory chemokine, IL-8 [41]. A recent study presented a genetic association of IL-8 polymorphisms with FEV1 status of CF patients [42]. In our study, we observed increased expression of IL-8 transcripts in individuals with mild CF. However, this observation could not be verified by qPCR as more IL-8 expression was found in the severe CF group. A similar results was observed in nasal epithelia cells between mild and severe CF and argued that IL-8 is not predictably associated with severity of the disease [17]. However, as we noted a contradicting result between RNA-seq and qPCR, further validation with independent samples seems essential to explore the role of IL-8 in CF lung function variability. GWAS meta-analysis study with a high sample number (n = 6,365) revealed the strong association of five genomic regions to CF lung disease severity [13]. We examined whether any of the observed DEG shares the specified genomic location. Remarkably, we found that chromosomal mapping has assigned the HCG27 gene to position 6p21.33, which is very close to the reported HLA-DRA locus at 6p21.32. Although HCG27 is reported to be a pseudo-gene, we observed a significant difference in expression levels between mild and severe CF individuals and the region including HCG27 is also a major susceptibility loci for psoriasis [43]. Nevertheless, our present study using a RNA-seq based transcriptomics approach was able to locate reported susceptibility loci and thereby replicate the larger GWAS study. Although other genes described earlier do not exactly match those identified in our study, they belong to the same biological families. For example, ATP12A was reported to be responsible for airway acidification in CF, and we identified ATP6V1G1, a gene of the same family, in our study [44].
Collectively, our comparative transcriptomic analysis between mild and severe CF lung disease provided new insights into CF pulmonary decline. The global gene expression analyses identified that genes of the type I interferon response and ribosomal stalk proteins and revealed potential CF modifier genes. Our findings have the potential for picking new therapeutic targets from from this list of genes for treatment of cystic fibrosis.
Supporting information
Absolute (A) and relative (B) number of reads mapped to hg19.
(TIFF)
PCA of the filtered CPM counts data to assess the differences in the expression profiles of the two investigated groups. Each point represents one sample; the closer two points are the more similar are the expression profiles. CF manifestation is color-coded.
(TIFF)
Network of MARA analysis predicts the possible interaction of IRF1,2,8 and STAT2 target genes.
(TIFF)
The results of differential expression analysis between mild and severe CF group. The respective expression levels given as a fold change and CPM with corrected P-Values.
(PDF)
The results of GO enrichement analysis for DEGs that are upregulated in mild and severe CF. Values of fold enrichment of specififed pathways with with corrected P-values were stated.
(PDF)
The complete raw data of MARA analysis that predicted the IRF1,2,8 and STAT2 activity in RNA-seq.
(PDF)
Acknowledgments
We thank Katrin Ganzenberg for help with proof-reading of the draft.
Data Availability
RNA-Seq reads are available in the NCBI-Sequence Read Archive (SRA) database, under accession number SRP111640.
Funding Statement
JSA was supported by Europe Research Council Starting Grant (to M.S.D.K., No. 637752). AD was supported by HMZ Private Foundation (to M.S.D.K.). The authors acknowledge the support of the Open Access Publishing Fund from University of Tübingen. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Elborn JS. Cystic fibrosis. Lancet. 2016. doi: 10.1016/S0140-6736(16)00576-6 . [DOI] [PubMed] [Google Scholar]
- 2.O'Sullivan BP, Freedman SD. Cystic fibrosis. Lancet. 2009;373(9678):1891–904. doi: 10.1016/S0140-6736(09)60327-5 . [DOI] [PubMed] [Google Scholar]
- 3.Cutting GR. Cystic fibrosis genetics: from molecular understanding to clinical application. Nat Rev Genet. 2015;16(1):45–56. doi: 10.1038/nrg3849 ; PubMed Central PMCID: PMC4364438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cai ZW, Liu J, Li HY, Sheppard DN. Targeting F508del-CFTR to develop rational new therapies for cystic fibrosis. Acta Pharmacol Sin. 2011;32(6):693–701. doi: 10.1038/aps.2011.71 ; PubMed Central PMCID: PMC4009972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wright FA, Strug LJ, Doshi VK, Commander CW, Blackman SM, Sun L, et al. Genome-wide association and linkage identify modifier loci of lung disease severity in cystic fibrosis at 11p13 and 20q13.2. Nat Genet. 2011;43(6):539–46. doi: 10.1038/ng.838 ; PubMed Central PMCID: PMC3296486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Correlation between genotype and phenotype in patients with cystic fibrosis. The Cystic Fibrosis Genotype-Phenotype Consortium. N Engl J Med. 1993;329(18):1308–13. doi: 10.1056/NEJM199310283291804 . [DOI] [PubMed] [Google Scholar]
- 7.Mekus F, Ballmann M, Bronsveld I, Bijman J, Veeze H, Tummler B. Categories of deltaF508 homozygous cystic fibrosis twin and sibling pairs with distinct phenotypic characteristics. Twin Res. 2000;3(4):277–93. . [DOI] [PubMed] [Google Scholar]
- 8.Vanscoy LL, Blackman SM, Collaco JM, Bowers A, Lai T, Naughton K, et al. Heritability of lung disease severity in cystic fibrosis. Am J Respir Crit Care Med. 2007;175(10):1036–43. doi: 10.1164/rccm.200608-1164OC ; PubMed Central PMCID: PMC1899267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cutting GR. Modifier genes in Mendelian disorders: the example of cystic fibrosis. Ann N Y Acad Sci. 2010;1214:57–69. doi: 10.1111/j.1749-6632.2010.05879.x ; PubMed Central PMCID: PMC3040597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hamilton BA, Yu BD. Modifier genes and the plasticity of genetic networks in mice. PLoS Genet. 2012;8(4):e1002644 doi: 10.1371/journal.pgen.1002644 ; PubMed Central PMCID: PMC3325199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Genin E, Feingold J, Clerget-Darpoux F. Identifying modifier genes of monogenic disease: strategies and difficulties. Hum Genet. 2008;124(4):357–68. doi: 10.1007/s00439-008-0560-2 ; PubMed Central PMCID: PMC2911473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clarke LA, Sousa L, Barreto C, Amaral MD. Changes in transcriptome of native nasal epithelium expressing F508del-CFTR and intersecting data from comparable studies. Respir Res. 2013;14:38 doi: 10.1186/1465-9921-14-38 ; PubMed Central PMCID: PMC3637641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Corvol H, Blackman SM, Boelle PY, Gallins PJ, Pace RG, Stonebraker JR, et al. Genome-wide association meta-analysis identifies five modifier loci of lung disease severity in cystic fibrosis. Nat Commun. 2015;6:8382 doi: 10.1038/ncomms9382 ; PubMed Central PMCID: PMC4589222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dorfman R, Sandford A, Taylor C, Huang B, Frangolias D, Wang Y, et al. Complex two-gene modulation of lung disease severity in children with cystic fibrosis. J Clin Invest. 2008;118(3):1040–9. doi: 10.1172/JCI33754 ; PubMed Central PMCID: PMC2248329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gu Y, Harley IT, Henderson LB, Aronow BJ, Vietor I, Huber LA, et al. Identification of IFRD1 as a modifier gene for cystic fibrosis lung disease. Nature. 2009;458(7241):1039–42. doi: 10.1038/nature07811 ; PubMed Central PMCID: PMC2841516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ogilvie V, Passmore M, Hyndman L, Jones L, Stevenson B, Wilson A, et al. Differential global gene expression in cystic fibrosis nasal and bronchial epithelium. Genomics. 2011;98(5):327–36. doi: 10.1016/j.ygeno.2011.06.008 . [DOI] [PubMed] [Google Scholar]
- 17.Wright JM, Merlo CA, Reynolds JB, Zeitlin PL, Garcia JG, Guggino WB, et al. Respiratory epithelial gene expression in patients with mild and severe cystic fibrosis lung disease. Am J Respir Cell Mol Biol. 2006;35(3):327–36. doi: 10.1165/rcmb.2005-0359OC ; PubMed Central PMCID: PMC2643286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao S, Fung-Leung WP, Bittner A, Ngo K, Liu X. Comparison of RNA-Seq and microarray in transcriptome profiling of activated T cells. PLoS One. 2014;9(1):e78644 doi: 10.1371/journal.pone.0078644 ; PubMed Central PMCID: PMC3894192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635 ; PubMed Central PMCID: PMC3530905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anders S, Pyl PT, Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–9. doi: 10.1093/bioinformatics/btu638 ; PubMed Central PMCID: PMC4287950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–40. doi: 10.1093/bioinformatics/btp616 ; PubMed Central PMCID: PMC2796818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30(9):e36 ; PubMed Central PMCID: PMC113859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gallati S. Disease-modifying genes and monogenic disorders: experience in cystic fibrosis. Appl Clin Genet. 2014;7:133–46. doi: 10.2147/TACG.S18675 ; PubMed Central PMCID: PMC4104546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang C, Gong B, Bushel PR, Thierry-Mieg J, Thierry-Mieg D, Xu J, et al. The concordance between RNA-seq and microarray data depends on chemical treatment and transcript abundance. Nat Biotechnol. 2014;32(9):926–32. doi: 10.1038/nbt.3001 ; PubMed Central PMCID: PMC4243706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Verhaeghe C, Remouchamps C, Hennuy B, Vanderplasschen A, Chariot A, Tabruyn SP, et al. Role of IKK and ERK pathways in intrinsic inflammation of cystic fibrosis airways. Biochem Pharmacol. 2007;73(12):1982–94. doi: 10.1016/j.bcp.2007.03.019 . [DOI] [PubMed] [Google Scholar]
- 26.Virella-Lowell I, Herlihy JD, Liu B, Lopez C, Cruz P, Muller C, et al. Effects of CFTR, interleukin-10, and Pseudomonas aeruginosa on gene expression profiles in a CF bronchial epithelial cell Line. Mol Ther. 2004;10(3):562–73. doi: 10.1016/j.ymthe.2004.06.215 . [DOI] [PubMed] [Google Scholar]
- 27.Zabner J, Scheetz TE, Almabrazi HG, Casavant TL, Huang J, Keshavjee S, et al. CFTR DeltaF508 mutation has minimal effect on the gene expression profile of differentiated human airway epithelia. Am J Physiol Lung Cell Mol Physiol. 2005;289(4):L545–53. doi: 10.1152/ajplung.00065.2005 . [DOI] [PubMed] [Google Scholar]
- 28.Trouve P, Genin E, Ferec C. In silico search for modifier genes associated with pancreatic and liver disease in Cystic Fibrosis. PLoS One. 2017;12(3):e0173822 doi: 10.1371/journal.pone.0173822 ; PubMed Central PMCID: PMC5365109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frickmann H, Jungblut S, Hirche TO, Gross U, Kuhns M, Zautner AE. Spectrum of viral infections in patients with cystic fibrosis. Eur J Microbiol Immunol (Bp). 2012;2(3):161–75. doi: 10.1556/EuJMI.2.2012.3.1 ; PubMed Central PMCID: PMC3962751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McNab F, Mayer-Barber K, Sher A, Wack A, O'Garra A. Type I interferons in infectious disease. Nat Rev Immunol. 2015;15(2):87–103. doi: 10.1038/nri3787 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smeekens SP, Ng A, Kumar V, Johnson MD, Plantinga TS, van Diemen C, et al. Functional genomics identifies type I interferon pathway as central for host defense against Candida albicans. Nat Commun. 2013;4:1342 doi: 10.1038/ncomms2343 ; PubMed Central PMCID: PMC3625375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parker D, Cohen TS, Alhede M, Harfenist BS, Martin FJ, Prince A. Induction of type I interferon signaling by Pseudomonas aeruginosa is diminished in cystic fibrosis epithelial cells. Am J Respir Cell Mol Biol. 2012;46(1):6–13. doi: 10.1165/rcmb.2011-0080OC ; PubMed Central PMCID: PMC3262660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blanchard AC, Horton E, Stanojevic S, Taylor L, Waters V, Ratjen F. Effectiveness of a stepwise Pseudomonas aeruginosa eradication protocol in children with cystic fibrosis. J Cyst Fibros. 2017. doi: 10.1016/j.jcf.2017.01.007 . [DOI] [PubMed] [Google Scholar]
- 34.Parker D, Martin FJ, Soong G, Harfenist BS, Aguilar JL, Ratner AJ, et al. Streptococcus pneumoniae DNA initiates type I interferon signaling in the respiratory tract. MBio. 2011;2(3):e00016–11. doi: 10.1128/mBio.00016-11 ; PubMed Central PMCID: PMC3101776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parker D, Prince A. Type I interferon response to extracellular bacteria in the airway epithelium. Trends Immunol. 2011;32(12):582–8. doi: 10.1016/j.it.2011.09.003 ; PubMed Central PMCID: PMC3221817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hector A, Kormann M, Kammermeier J, Burdi S, Marcos V, Rieber N, et al. Expression and regulation of interferon-related development regulator-1 in cystic fibrosis neutrophils. Am J Respir Cell Mol Biol. 2013;48(1):71–7. doi: 10.1165/rcmb.2012-0061OC . [DOI] [PubMed] [Google Scholar]
- 37.Balloy V, Varet H, Dillies MA, Proux C, Jagla B, Coppee JY, et al. Normal and Cystic Fibrosis Human Bronchial Epithelial Cells Infected with Pseudomonas aeruginosa Exhibit Distinct Gene Activation Patterns. PLoS One. 2015;10(10):e0140979 doi: 10.1371/journal.pone.0140979 ; PubMed Central PMCID: PMC4618526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen F, Zhang C, Jia X, Wang S, Wang J, Chen Y, et al. Transcriptome Profiles of Human Lung Epithelial Cells A549 Interacting with Aspergillus fumigatus by RNA-Seq. PLoS One. 2015;10(8):e0135720 doi: 10.1371/journal.pone.0135720 ; PubMed Central PMCID: PMC4537115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Veit G, Oliver K, Apaja PM, Perdomo D, Bidaud-Meynard A, Lin ST, et al. Ribosomal Stalk Protein Silencing Partially Corrects the DeltaF508-CFTR Functional Expression Defect. PLoS Biol. 2016;14(5):e1002462 doi: 10.1371/journal.pbio.1002462 ; PubMed Central PMCID: PMC4864299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Valdivieso AG, Clauzure M, Massip-Copiz M, Santa-Coloma TA. The Chloride Anion Acts as a Second Messenger in Mammalian Cells—Modifying the Expression of Specific Genes. Cell Physiol Biochem. 2016;38(1):49–64. doi: 10.1159/000438608 . [DOI] [PubMed] [Google Scholar]
- 41.Jundi K, Greene CM. Transcription of Interleukin-8: How Altered Regulation Can Affect Cystic Fibrosis Lung Disease. Biomolecules. 2015;5(3):1386–98. doi: 10.3390/biom5031386 ; PubMed Central PMCID: PMC4598756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Furlan LL, Marson FA, Ribeiro JD, Bertuzzo CS, Salomao Junior JB, Souza DR. IL8 gene as modifier of cystic fibrosis: unraveling the factors which influence clinical variability. Hum Genet. 2016;135(8):881–94. doi: 10.1007/s00439-016-1684-4 . [DOI] [PubMed] [Google Scholar]
- 43.Nair RP, Stuart PE, Nistor I, Hiremagalore R, Chia NV, Jenisch S, et al. Sequence and haplotype analysis supports HLA-C as the psoriasis susceptibility 1 gene. Am J Hum Genet. 2006;78(5):827–51. doi: 10.1086/503821 ; PubMed Central PMCID: PMC1474031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shah VS, Meyerholz DK, Tang XX, Reznikov L, Abou Alaiwa M, Ernst SE, et al. Airway acidification initiates host defense abnormalities in cystic fibrosis mice. Science. 2016;351(6272):503–7. doi: 10.1126/science.aad5589 ; PubMed Central PMCID: PMC4852973. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Absolute (A) and relative (B) number of reads mapped to hg19.
(TIFF)
PCA of the filtered CPM counts data to assess the differences in the expression profiles of the two investigated groups. Each point represents one sample; the closer two points are the more similar are the expression profiles. CF manifestation is color-coded.
(TIFF)
Network of MARA analysis predicts the possible interaction of IRF1,2,8 and STAT2 target genes.
(TIFF)
The results of differential expression analysis between mild and severe CF group. The respective expression levels given as a fold change and CPM with corrected P-Values.
(PDF)
The results of GO enrichement analysis for DEGs that are upregulated in mild and severe CF. Values of fold enrichment of specififed pathways with with corrected P-values were stated.
(PDF)
The complete raw data of MARA analysis that predicted the IRF1,2,8 and STAT2 activity in RNA-seq.
(PDF)
Data Availability Statement
RNA-Seq reads are available in the NCBI-Sequence Read Archive (SRA) database, under accession number SRP111640.