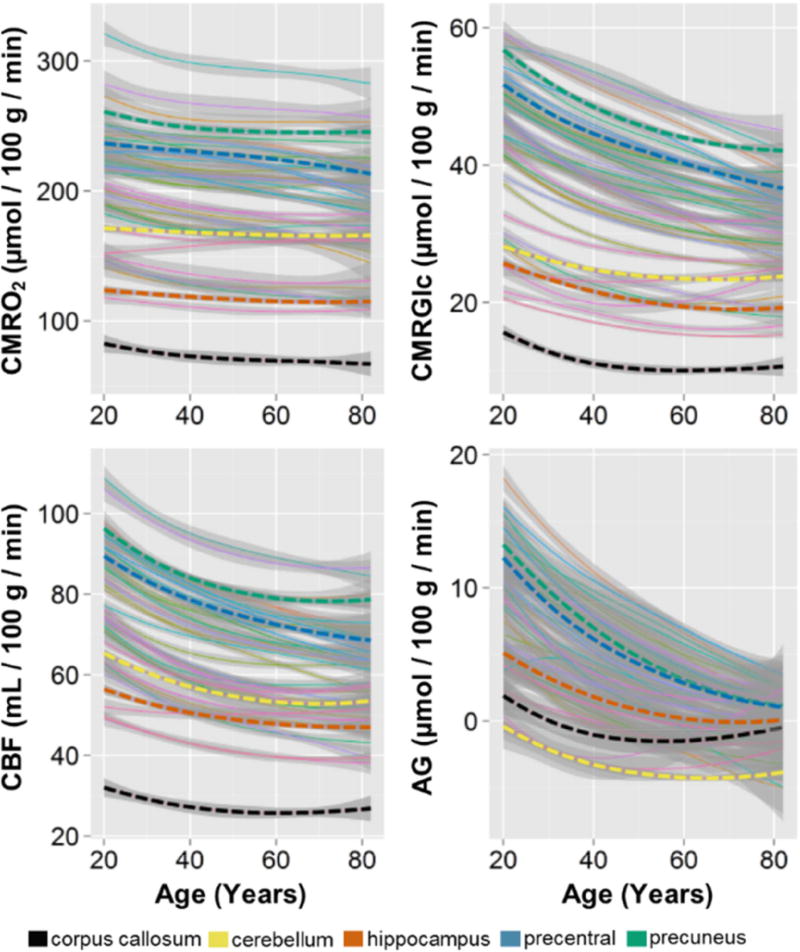
Figure 3. Summary of regional quantitative metabolic change in the human brain with aging.
Literature-based whole brain estimates for CMRO2, CMRGlc, CBF, and AG were combined with local-to-global ratios to determine a quantitative value in every individual in the normative cohort, for each metabolic parameter in 42 gray matter regions and the corpus callosum as a representative white matter region. These were then fit with loess curves to demonstrate the trajectory for each metabolic parameter in each of these regions. The thin lines represent each of the regions with superimposed thick dotted lines representing representative regions, as shown in the bottom legend. The shaded gray regions show the standard error about the loess curves, though it should be noted that using literature-based whole brain estimates will necessarily obscure quantitative inter-individual variability. It is evident that with rare exception, all regions decrease in all aspects of metabolism. However, the loss of CMRO2 is slight as compared to the more dramatic decrease in AG, which regionally varies in rate of change with aging. This whole-brain normalized regionally quantitative data is provided as a normative data set in the Supplemental Table.