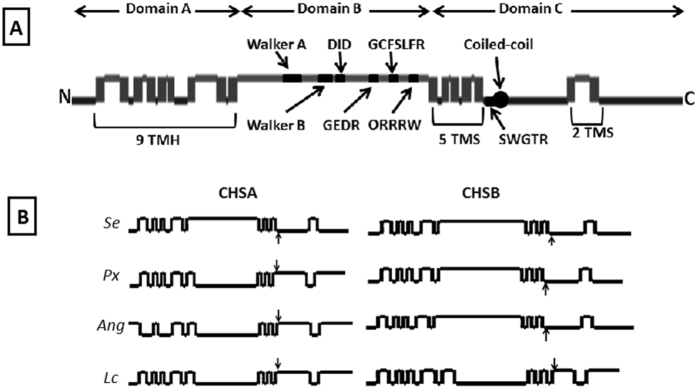
Figure 2.
Predicted structure of PhoCHSA and selected CHS enzymes identified from other insects using TMHMM-2.0. (A) Topology of PhoCHSA showing the three major structural domains and associated structures and motifs. TMH: transmembrane helix, TMS: transmembrane span, N: N-terminus and C: C-terminus. Vertical bars represent the transmembrane regions while horizontal lines represent inner (lower) and outer (upper) domains. (B) Topology of selected CHS proteins showing the location of the catalytic domain and the SWGTR motif (indicated by arrow). Catalytic domains of SeCHSA (AAZ03545), SeCHSB (ABI96087), PxCHSB (XP_011565203) and AngCHSB (AGAP001205-PA) are located outside the plasma membrane while catalytic domains of PxCHSA (BAF47974), AngCHSA (AGAP001748), LcCHSA (AAG09712) and LcCHSB (KNC29062) are located inside the plasma membrane and facing the cytoplasm. Catalytic domain and SWGTR motif are always on opposite sides of the plasma membrane. Ang: Anopheles gambiae, Lc: Lucilia cuprina, Px: Plutella xylostella, Se: Spodoptera exigua.