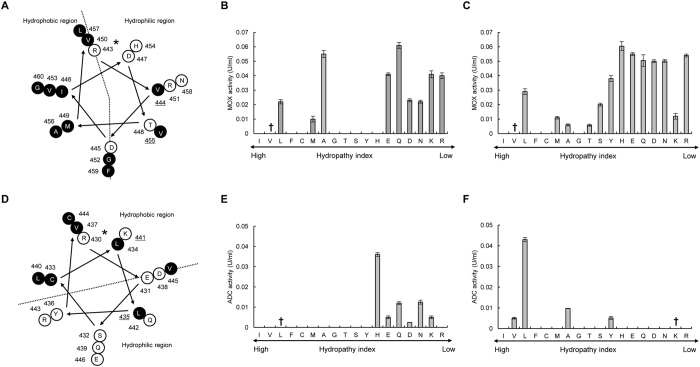
Figure 1.
Helical wheel depictions for α-helix regions of four enzymes that contributed to improving the protein solubility, and saturation mutagenesis at aggregation hotspots on α-helices of ChMOX and AtADC identified by directed evolution analysis. Helical wheels depict the following α-helix regions: residues 443–460 (RVDIDTMVRGVHVALNFG) of ChMOX (A) and residues 430–446 (RESCLLYVDQLKQRCVE) of AtADC (D). Hydrophobic and hydrophilic residues are shown in white and black letters, respectively. The mutation sites for saturation mutagenesis are represented by underlined residue numbers. An asterisk (*) adjacent to the sequence number indicates the first residue of an α-helix. The enzyme activity of the saturated mutants was measured for the following four residues: V455X (B) and V444X (C) for ChMOX and K441X (E) and L435X (F) for AtADC. The residues with low and high hydropathy indices are hydrophilic and hydrophobic, respectively. The dagger (†) indicates WT ChMOX (B,C) and AtADC (E,F). Trp and Pro are not shown in these figures because the side chains of these residues exhibit hydrophobic character despite being classified as hydrophilic groups in the hydropathy index.