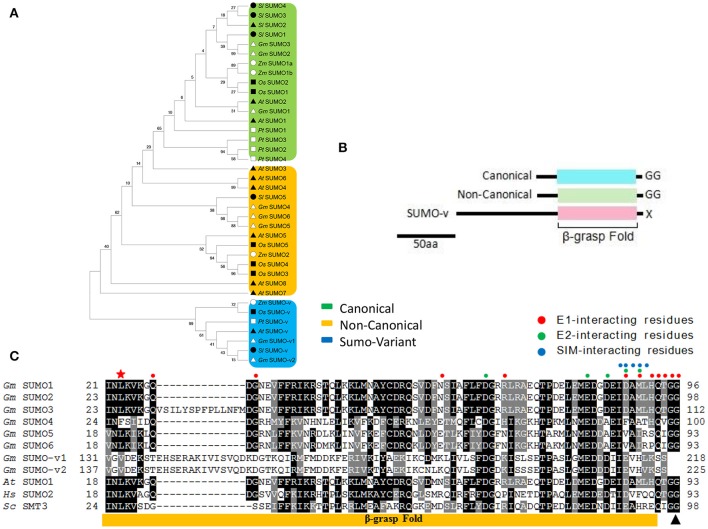
Figure 1.
The genomes of soybean and other plant species encode a family of SUMO-related proteins (A) Protein sequences from Arabidopsis thaliana, Zea mays, Oryza sativa, Solanum lycopersium, Populus trichocarpa and Glycine max (Table S2) were used to construct the phylogenetic tree by the neighbor-joining method in MEGA. They were classified into three groups: canonical SUMO, non-canonical SUMO, and SUMO-variant. (B) Domain structures of the plant SUMO family that include the canonical and non-canonical SUMO isoforms, and SUMO-variant containing a long, conserved N-terminal extension in front of the β-grasp domain. (C) Alignment of SUMO sequence reveals conserved and divergent residues. Included are soybean (Gm) SUMO-1, -2, -3, -4, -5, -6, and SUMO-v1, -v2, along with canonical SUMOs from Arabidopsis (AtSUMO1), human (HsSUMO2, NP_008868.3), and yeast (ScSmt3, KZV12750.1), only conserved region is shown in the figure. The yellow box locates the β-grasp-fold. The black triangle locates the processing site by ULP that exposes the diGly motif essential for conjugation in canonical SUMOs. Residues marked with red, green, and blue circle dots are important for the non-covalent binding of SUMOs to E1, E2, and SIMs (SUMO interacting motif), respectively. The asterisk denotes the conserved Lys required for forming SUMO-chains. Gray and black boxes identify similar and conserved amino acids, respectively. Dashes denote gaps.